Escherichia coli genes affecting recipient ability in plasmid conjugation: are there any?
- PMID: 19203375
- PMCID: PMC2645431
- DOI: 10.1186/1471-2164-10-71
Escherichia coli genes affecting recipient ability in plasmid conjugation: are there any?
Abstract
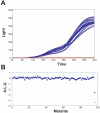
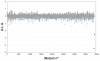
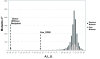
Background: How does the recipient cell contribute to bacterial conjugation? To answer this question we systematically analyzed the individual contribution of each Escherichia coli gene in matings using plasmid R388 as a conjugative plasmid. We used an automated conjugation assay and two sets of E. coli mutant collections: the Keio collection (3,908 E. coli single-gene deletion mutants) and a collection of 20,000 random mini-Tn10::Km insertion mutants in E. coli strain DH5alpha. The combined use of both collections assured that we screened > 99% of the E. coli non-essential genes in our survey.
Results: Results indicate that no non-essential recipient E. coli genes exist that play an essential role in conjugation. Mutations in the lipopolysaccharide (LPS) synthesis pathway had a modest effect on R388 plasmid transfer (6 - 32% of wild type). The same mutations showed a drastic inhibition effect on F-plasmid transfer, but only in liquid matings, suggesting that previously isolated conjugation-defective mutants do in fact impair mating pair formation in liquid mating, but not conjugative DNA processing or transport per se.
Conclusion: We conclude from our genome-wide screen that recipient bacterial cells cannot avoid being used as recipients in bacterial conjugation. This is relevant as an indication of the problems in curbing the dissemination of antibiotic resistance and suggests that conjugation acts as a pure drilling machine, with little regard to the constitution of the recipient cell.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

