Regulation of nuclear import and export of negative cofactor 2
- PMID: 19204005
- PMCID: PMC2666590
- DOI: 10.1074/jbc.M805694200
Regulation of nuclear import and export of negative cofactor 2
Abstract
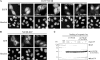
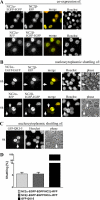
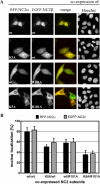
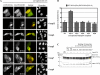
The negative cofactor 2 (NC2) is a protein complex composed of two subunits, NC2alpha and NC2beta, and plays a key role in transcription regulation. Here we investigate whether each subunit contains a nuclear localization signal (NLS) that permits individual crossing of the nuclear membrane or whether nuclear import of NC2alpha and NC2beta depends on heterodimerization. Our results from in vitro binding studies and transfection experiments in cultured cells show that each subunit contains a classical NLS (cNLS) that is recognized by the importin alpha/beta heterodimer. Regardless of the individual cNLSs the two NC2 subunits are translocated as a preassembled complex as co-transfection experiments with wild-type and cNLS-deficient NC2 subunits demonstrate. Ran-dependent binding of the nuclear export receptor Crm1/exportin 1 confirmed the presence of a leucine-rich nuclear export signal (NES) in NC2beta. In contrast, NC2alpha does not exhibit a NES. Our results from interspecies heterokaryon assays suggest that heterodimerization with NC2alpha masks the NES in NC2beta, which prevents nuclear export of the NC2 complex. A mutation in either one of the two cNLSs decreases the extent of importin alpha/beta-mediated nuclear import of the NC2 complex. In addition, the NC2 complex can enter the nucleus via a second pathway, facilitated by importin 13. Because importin 13 binds exclusively to the NC2 complex but not to the individual subunits this alternative import pathway depends on sequence elements distributed among the two subunits.
Figures


Similar articles
-
Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-beta.J Mol Biol. 1997 Dec 19;274(5):693-707. doi: 10.1006/jmbi.1997.1420. J Mol Biol. 1997. PMID: 9405152
-
Quantification of nuclear protein transport using induced heterodimerization.Traffic. 2009 Sep;10(9):1221-7. doi: 10.1111/j.1600-0854.2009.00953.x. Epub 2009 Jun 9. Traffic. 2009. PMID: 19602195
-
Identification of nuclear localization signal and nuclear export signal of VP1 from the chicken anemia virus and effects on VP2 shuttling in cells.Virol J. 2019 Apr 5;16(1):45. doi: 10.1186/s12985-019-1153-5. Virol J. 2019. PMID: 30953524 Free PMC article.
-
Nucleocytoplasmic protein transport and recycling of Ran.Cell Struct Funct. 1999 Dec;24(6):425-33. doi: 10.1247/csf.24.425. Cell Struct Funct. 1999. PMID: 10698256 Review.
-
The nucleocytoplasmic transport of viral proteins.Virol Sin. 2010 Apr;25(2):79-85. doi: 10.1007/s12250-010-3099-z. Epub 2010 Apr 9. Virol Sin. 2010. PMID: 20960304 Free PMC article. Review.
Cited by
-
Several phenylalanine-glycine motives in the nucleoporin Nup214 are essential for binding of the nuclear export receptor CRM1.J Biol Chem. 2013 Feb 8;288(6):3952-63. doi: 10.1074/jbc.M112.433243. Epub 2012 Dec 21. J Biol Chem. 2013. PMID: 23264634 Free PMC article.
-
Importin 13 mediates nuclear import of histone fold-containing chromatin accessibility complex heterodimers.J Biol Chem. 2009 Apr 24;284(17):11652-62. doi: 10.1074/jbc.M806820200. Epub 2009 Feb 14. J Biol Chem. 2009. PMID: 19218565 Free PMC article.
-
Physiological change under OsHV-1 contamination in Pacific oyster Crassostrea gigas through massive mortality events on fields.BMC Genomics. 2013 Aug 29;14:590. doi: 10.1186/1471-2164-14-590. BMC Genomics. 2013. PMID: 23987141 Free PMC article.
-
Nuclear transport signals control cellular localization and function of androgen receptor cofactor p44/WDR77.PLoS One. 2011;6(7):e22395. doi: 10.1371/journal.pone.0022395. Epub 2011 Jul 15. PLoS One. 2011. PMID: 21789256 Free PMC article.
-
The anti-inflammatory prostaglandin 15-deoxy-delta(12,14)-PGJ2 inhibits CRM1-dependent nuclear protein export.J Biol Chem. 2010 Jul 16;285(29):22202-10. doi: 10.1074/jbc.M110.131821. Epub 2010 May 10. J Biol Chem. 2010. PMID: 20457605 Free PMC article.
References
-
- Mermelstein, F., Yeung, K., Cao, J., Inostroza, J. A., Erdjument-Bromage, H., Eagelson, K., Landsman, D., Levitt, P., Tempst, P., and Reinberg, D. (1996) Genes Dev. 10 1033-1048 - PubMed
-
- Kamada, K., Shu, F., Chen, H., Malik, S., Stelzer, G., Roeder, R. G., Meisterernst, M., and Burley, S. K. (2001) Cell 106 71-81 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

