Functional characterization of two secreted SEL1L isoforms capable of exporting unassembled substrate
- PMID: 19204006
- PMCID: PMC2670146
- DOI: 10.1074/jbc.M805408200
Functional characterization of two secreted SEL1L isoforms capable of exporting unassembled substrate
Abstract
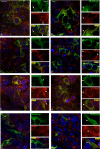
SEL1L-A, a transmembrane glycoprotein residing in the endoplasmic reticulum (ER), is a component of the ER-associated degradation (ERAD) pathway. Alternative splicing generates two smaller SEL1L isoforms, -B and -C, that lack the SEL1L-A membrane-spanning region but retain some sel-1-like repeats, known to be involved in multi-protein interactions and signal transduction. In this study the functional characteristics of SEL1L-B and -C were investigated in human cell models. We show that these two isoforms are induced upon ER stress and activation of the unfolded protein response, together with SEL1L-A. Using transient transfection experiments (based on wild-type and mutant SEL1L constructs) combined with several biochemical tests we show that SEL1L-B and, more prominently, SEL1L-C are secreted glycoproteins. Although SEL1L-C is in monomeric form, SEL1L-B is engaged in intramolecular/intermolecular disulfide bonds. Both isoforms localize in secretory and degradative cellular compartments and in areas of cell-cell contact. However, whereas SEL1L-B is mainly associated with membranes, SEL1L-C shows the typical intralumenal localization of soluble proteins and is present in intercellular spaces. Furthermore, because of its peroxisomal domain, SEL1L-C localizes to peroxisomes. Both SEL1L-B and -C are involved in sorting and exporting unassembled Ig-mu(s) but do not affect two other ERAD substrates, the null Hong Kong variant of alpha(1)-antitrypsin, and mutant alpha(1)-AT Z. Overall these findings suggest that SEL1L-B and -C participate to novel molecular pathways that, in parallel with ERAD, contribute to the disposure of misfolded/unfolded or orphan proteins through degradation or secretion.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

