Founders, drift, and infidelity: the relationship between Y chromosome diversity and patrilineal surnames
- PMID: 19204044
- PMCID: PMC2668828
- DOI: 10.1093/molbev/msp022
Founders, drift, and infidelity: the relationship between Y chromosome diversity and patrilineal surnames
Abstract
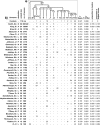
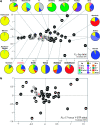
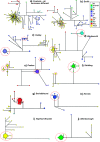
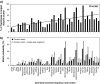
Most heritable surnames, like Y chromosomes, are passed from father to son. These unique cultural markers of coancestry might therefore have a genetic correlate in shared Y chromosome types among men sharing surnames, although the link could be affected by mutation, multiple foundation for names, nonpaternity, and genetic drift. Here, we demonstrate through an analysis of 1,678 Y-chromosomal haplotypes within 40 British surnames a remarkably high degree of coancestry that generally increases as surnames become rarer. On average, the proportion of haplotypes lying within descent clusters is 62% but ranges from 0% to 87%. The shallow time depth of many descent clusters within names, the lack of a detectable effect of surname derivation on diversity, and simulations of surname descent suggest that genetic drift through variation in reproductive success is important in structuring haplotype diversity. Modern patterns therefore provide little reliable information about the original founders of surnames some 700 years ago. A comparative analysis of published data on Y diversity within Irish surnames demonstrates a relative lack of surname frequency dependence of coancestry, a difference probably mediated through distinct Irish and British demographic histories including even more marked genetic drift in Ireland.
Figures
References
-
- Adams SM, King TE, Bosch E, Jobling MA. The case of the unreliable SNP: recurrent back-mutation of Y-chromosomal marker P25 through gene conversion. Forensic Sci Int. 2006;159:14–20. - PubMed
-
- Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. - PubMed
-
- Bosch E, Calafell F, González-Neira A, et al. Male and female lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann Hum Genet. 2006;70:459–487. - PubMed
-
- Bosch E, Lee AC, Calafell F, Arroyo E, Henneman P, de Knijff P, Jobling MA. High resolution Y chromosome typing: 19 STRs amplified in three multiplex reactions. Forensic Sci Int. 2002;125:42–51. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

