Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III
- PMID: 19204085
- PMCID: PMC2663309
- DOI: 10.1128/MCB.01841-08
Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III
Abstract
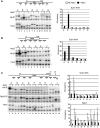
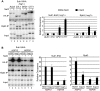
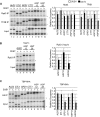
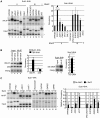
Sub1 is implicated in transcriptional activation, elongation, and mRNA 3'-end formation in budding yeast. To gain more insight into its function, we performed a synthetic genetic array screen with SUB1 that uncovered genetic interactions with genes involved in the high-osmolarity glycerol (HOG) osmoresponse pathway. We find that Sub1 and the HOG pathway are redundant for survival in moderate osmolarity. Chromatin immunoprecipitation analysis shows that Sub1 is recruited to osmoresponse gene promoters during osmotic shock and is required for full recruitment of TBP, TFIIB, and RNA polymerase II (RNAP II) at a subset of these genes. Furthermore, we detect Sub1 at the promoter of every constitutively transcribed RNAP II and, unexpectedly, at every RNAP III gene tested, but not at the RNAP I-transcribed ribosomal DNA promoter. Significantly, deletion of SUB1 reduced levels of promoter-associated RNAP II or III at these genes, but not TBP levels. Together these data suggest that, in addition to a general role in polymerase recruitment at constitutive RNAP II and RNAP III genes, during osmotic shock, Sub1 facilitates osmoresponse gene transcription by enhancing preinitiation complex formation.
Figures



References
-
- Albertyn, J., S. Hohmann, J. M. Thevelein, and B. A. Prior. 1994. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 144135-4144. - PMC - PubMed
-
- Alepuz, P. M., A. Jovanovic, V. Reiser, and G. Ammerer. 2001. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell 7767-777. - PubMed
-
- Amberg, D. C., D. J. Burke, and J. N. Strathern. 2006. Yeast RNA isolation: small-scale. Cold Spring Harbor protocol. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. doi: 10.1101/pdb.prot4155. - DOI
-
- Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 2591760-1763. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
