Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola
- PMID: 19204088
- PMCID: PMC2663137
- DOI: 10.1128/IAI.01544-08
Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola
Abstract
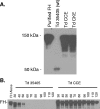
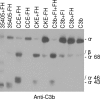
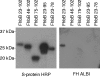
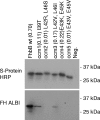
Treponema denticola, a spirochete associated with periodontitis, is abundant at the leading edge of subgingival plaque, where it interacts with gingival epithelia. T. denticola produces a number of virulence factors, including dentilisin, a protease which is cytopathic to host cells, and FhbB, a unique T. denticola lipoprotein that binds complement regulatory proteins. Earlier analyses suggested that FhbB specifically bound to factor H (FH)-like protein 1 (FHL-1). However, by using dentilisin-deficient mutants of T. denticola, we found that T. denticola preferentially binds FH and not FHL-1, and that FH is then cleaved by dentilisin to yield an FH subfragment of approximately 50 kDa. FH bound to dentilisin-deficient mutants but was not cleaved and retained its ability to serve as a cofactor for factor I in the cleavage of C3b. To assess the molecular basis of the interaction of FhbB with FH, mutational analyses were conducted. Replacement of specific residues in widely separated domains of FhbB and disruption of a central alpha helix with coiled-coil formation probability attenuated or eliminated FH binding. The data presented here are the first to demonstrate the retention at the cell surface of a proteolytic cleavage product of FH. The precise role of this FH fragment in the host-pathogen interaction remains to be determined.
Figures



Similar articles
-
Identification of the primary mechanism of complement evasion by the periodontal pathogen, Treponema denticola.Mol Oral Microbiol. 2011 Apr;26(2):140-9. doi: 10.1111/j.2041-1014.2010.00598.x. Epub 2010 Dec 3. Mol Oral Microbiol. 2011. PMID: 21375704 Free PMC article.
-
Analysis of the complement sensitivity of oral treponemes and the potential influence of FH binding, FH cleavage and dentilisin activity on the pathogenesis of periodontal disease.Mol Oral Microbiol. 2014 Oct;29(5):194-207. doi: 10.1111/omi.12054. Epub 2014 Jun 3. Mol Oral Microbiol. 2014. PMID: 24815960 Free PMC article.
-
Plasminogen binding and degradation by Treponema denticola: Identification of the plasminogen binding interface on the FhbB protein.Mol Oral Microbiol. 2018 Jun;33(3):249-256. doi: 10.1111/omi.12221. Epub 2018 Apr 23. Mol Oral Microbiol. 2018. PMID: 29498487
-
Approaches to Understanding Mechanisms of Dentilisin Protease Complex Expression in Treponema denticola.Front Cell Infect Microbiol. 2021 May 18;11:668287. doi: 10.3389/fcimb.2021.668287. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34084756 Free PMC article. Review.
-
Treponema denticola major surface protein (Msp): a key player in periodontal pathogenicity and immune evasion.Arch Microbiol. 2025 Jan 18;207(2):36. doi: 10.1007/s00203-024-04223-w. Arch Microbiol. 2025. PMID: 39825920 Review.
Cited by
-
Dentipain, a Streptococcus pyogenes IdeS protease homolog, is a novel virulence factor of Treponema denticola.Biol Chem. 2010 Sep;391(9):1047-55. doi: 10.1515/BC.2010.113. Biol Chem. 2010. PMID: 20635859 Free PMC article.
-
Hijacking Factor H for Complement Immune Evasion.Front Immunol. 2021 Feb 25;12:602277. doi: 10.3389/fimmu.2021.602277. eCollection 2021. Front Immunol. 2021. PMID: 33717083 Free PMC article. Review.
-
Treponema denticola chymotrypsin-like proteinase (CTLP) integrates spirochaetes within oral microbial communities.Microbiology (Reading). 2012 Mar;158(Pt 3):759-770. doi: 10.1099/mic.0.055939-0. Epub 2012 Feb 7. Microbiology (Reading). 2012. PMID: 22313692 Free PMC article.
-
The outer membrane protease PgtE of Salmonella enterica interferes with the alternative complement pathway by cleaving factors B and H.Front Microbiol. 2015 Feb 6;6:63. doi: 10.3389/fmicb.2015.00063. eCollection 2015. Front Microbiol. 2015. PMID: 25705210 Free PMC article.
-
Treponema denticola PrcB is required for expression and activity of the PrcA-PrtP (dentilisin) complex.J Bacteriol. 2010 Jul;192(13):3337-44. doi: 10.1128/JB.00274-10. Epub 2010 Apr 30. J Bacteriol. 2010. PMID: 20435733 Free PMC article.
References
-
- Alitalo, A., T. Meri, T. Chen, H. Lankinen, Z. Z. Cheng, T. S. Jokiranta, I. J. Seppala, P. Lahdenne, P. S. Hefty, D. R. Akins, and S. Meri. 2004. Lysine-dependent multipoint binding of the Borrelia burgdorferi virulence factor outer surface protein E to the C terminus of factor H. J. Immunol. 1726195-6201. - PubMed
-
- Alitalo, A., T. Meri, H. Lankinen, I. Seppala, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 1693847-3853. - PubMed
-
- Anonymous. 1987. Oral health of US adults: NIDR 1985 national survey. J. Public Health Dent. 47198-205. - PubMed
-
- Areschoug, T., M. Stalhammar-Carlemalm, I. Karlsosson, and G. Lindahl. 2002. Streptococcal beta protein has separate binding sites for human factor H and IgA-Fc. J. Biol. Chem. 27712642-12648. - PubMed
-
- Asakawa, R., H. Komatsuzawa, T. Kawai, S. Yamada, R. B. Goncalves, S. Izumi, T. Fujiwara, Y. Nakano, H. Shiba, M. A. Taubman, H. Kurihara, and M. Sugai. 2003. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 501125-1139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

