A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair
- PMID: 19204116
- PMCID: PMC2648549
- DOI: 10.1101/gad.1751209
A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair
Abstract
A DNA double-strand break (DSB) is repaired by gene conversion (GC) if both ends of the DSB share homology with an intact DNA sequence. However, if homology is limited to only one of the DSB ends, repair occurs by break-induced replication (BIR). It is not known how the homology status of the DSB ends is first assessed and what other parameters govern the choice between these repair pathways. Our data suggest that a "recombination execution checkpoint" (REC) regulates the choice of the homologous recombination pathway employed to repair a given DSB. This choice is made prior to the initiation of DNA synthesis, and is dependent on the relative position and orientation of the homologous sequences used for repair. The RecQ family helicase Sgs1 plays a key role in regulating the choice of the recombination pathway. Surprisingly, break repair and gap repair are fundamentally different processes, both kinetically and genetically, as Pol32 is required only for gap repair. We propose that the REC may have evolved to preserve genome integrity by promoting conservative repair, especially when a DSB occurs within a repeated sequence.
Figures
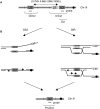
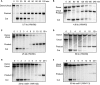

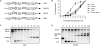

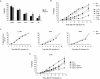

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
