Crystal structure of the eIF4A-PDCD4 complex
- PMID: 19204291
- PMCID: PMC2651268
- DOI: 10.1073/pnas.0808275106
Crystal structure of the eIF4A-PDCD4 complex
Abstract
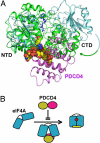
Tumor suppressor programmed cell death protein 4 (PDCD4) inhibits the translation initiation factor eIF4A, an RNA helicase that catalyzes the unwinding of secondary structure at the 5'-untranslated region of mRNAs and controls the initiation of translation. Here, we determined the crystal structure of the human eIF4A and PDCD4 complex. The structure reveals that one molecule of PDCD4 binds to the two eIF4A molecules through the two different binding modes. While the two MA3 domains of PDCD4 bind to one eIF4A molecule, the C-terminal MA3 domain alone of the same PDCD4 also interacts with another eIF4A molecule. The eIF4A-PDCD4 complex structure suggests that the MA3 domain(s) of PDCD4 binds perpendicular to the interface of the two domains of eIF4A, preventing the domain closure of eIF4A and blocking the binding of RNA to eIF4A, both of which are required events in the function of eIF4A helicase. The structure, together with biochemical analyses, reveals insights into the inhibition mechanism of eIF4A by PDCD4 and provides a framework for designing chemicals that target eIF4A.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
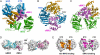

References
-
- Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65:6034–6041. - PubMed
-
- Chen Y, et al. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol. 2003;200:640–646. - PubMed
-
- Yang HS, Knies J, Stark C, Colburn NH. PDCD4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene. 2003;22:3712–3720. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

