Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata
- PMID: 19204294
- PMCID: PMC2637908
- DOI: 10.1073/pnas.0809793106
Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata
Abstract
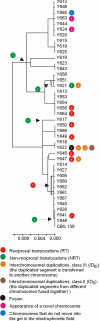
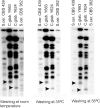
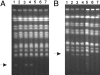
In eukaryotes, the number and rough organization of chromosomes is well preserved within isolates of the same species. Novel chromosomes and loss of chromosomes are infrequent and usually associated with pathological events. Here, we analyzed 40 pathogenic isolates of a haploid and asexual yeast, Candida glabrata, for their genome structure and stability. This organism has recently become the second most prevalent yeast pathogen in humans. Although the gene sequences were well conserved among different strains, their chromosome structures differed drastically. The most frequent events reshaping chromosomes were translocations of chromosomal arms. However, also larger segmental duplications were frequent and occasionally we observed novel chromosomes. Apparently, this yeast can generate a new chromosome by duplication of chromosome segments carrying a centromere and subsequently adding novel telomeric ends. We show that the observed genome plasticity is connected with antifungal drug resistance and it is likely an advantage in the human body, where environmental conditions fluctuate a lot.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Gauwerky CE, Croce CM. Chromosomal translocations in leukemia. Semin Cancer Biol. 1993;4:333–340. - PubMed
-
- Sharpless NE, et al. Impaired nonhomologous end-joining provokes soft tissue sarcomas harboring chromosomal translocations, amplifications, and deletions. Mol Cell. 2001;8:1187–1196. - PubMed
-
- Delneri D, et al. Engineering evolution to study speciation in yeasts. Nature. 2003;422:68–72. - PubMed
-
- Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. - PubMed
-
- Klempp-Selb B, Rimek D, Kappe R. Karyotyping of Candida albicans and Candida glabrata from patients with Candida sepsis. Mycoses. 2000;43:159–163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

