The molecular signature of CD8+ T cells undergoing deletional tolerance
- PMID: 19204323
- PMCID: PMC2680364
- DOI: 10.1182/blood-2008-10-185223
The molecular signature of CD8+ T cells undergoing deletional tolerance
Abstract
Peripheral tolerance induction is critical for the maintenance of self-tolerance and can be mediated by immunoregulatory T cells or by direct induction of T-cell anergy or deletion. Although the molecular processes underlying anergy have been extensively studied, little is known about the molecular basis for peripheral T-cell deletion. Here, we determined the gene expression signature of peripheral CD8(+) T cells undergoing deletional tolerance, relative to those undergoing immunogenic priming or lymphopenia-induced proliferation. From these data, we report the first detailed molecular signature of cells undergoing deletion. Consistent with defective cytolysis, these cells exhibited deficiencies in granzyme up-regulation. Furthermore, they showed antigen-driven Bcl-2 down-regulation and early up-regulation of the proapoptotic protein Bim, consistent with the requirement of this BH3-only protein for peripheral T-cell deletion. Bim up-regulation was paralleled by defective interleukin-7 receptor alpha (IL-7Ralpha) chain reexpression, suggesting that Bim-dependent death may be triggered by loss of IL-7/IL-7R signaling. Finally, we observed parallels in molecular signatures between deletion and anergy, suggesting that these tolerance pathways may not be as molecularly distinct as previously surmised.
Figures

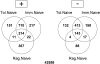

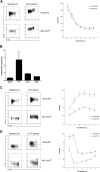
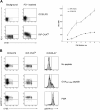
References
-
- Redmond WL, Sherman LA. Peripheral tolerance of CD8 T lymphocytes. Immunity. 2005;22:275–284. - PubMed
-
- Schwartz RH. T cell anergy. Annu Rev Immunol. 2003;21:305–334. - PubMed
-
- Redmond WL, Marincek BC, Sherman LA. Distinct requirements for deletion versus anergy during CD8 T cell peripheral tolerance in vivo. J Immunol. 2005;174:2046–2053. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

