Nucleosome positioning and gene regulation: advances through genomics
- PMID: 19204718
- PMCID: PMC4860946
- DOI: 10.1038/nrg2522
Nucleosome positioning and gene regulation: advances through genomics
Abstract
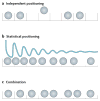
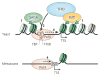
Knowing the precise locations of nucleosomes in a genome is key to understanding how genes are regulated. Recent 'next generation' ChIP-chip and ChIP-Seq technologies have accelerated our understanding of the basic principles of chromatin organization. Here we discuss what high-resolution genome-wide maps of nucleosome positions have taught us about how nucleosome positioning demarcates promoter regions and transcriptional start sites, and how the composition and structure of promoter nucleosomes facilitate or inhibit transcription. A detailed picture is starting to emerge of how diverse factors, including underlying DNA sequences and chromatin remodelling complexes, influence nucleosome positioning.
Figures
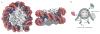
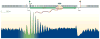

References
-
- Kornberg RD, Klug A. The nucleosome. Sci Am. 1981;244:52–64. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. - PubMed
-
- Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19:295–310. - PubMed
-
- Sarma K, Reinberg D. Histone variants meet their match. Nature Rev Mol Cell Biol. 2005;6:139–149. - PubMed
-
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature Genet. 2004;36:900–905. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

