Slow-onset inhibition of the FabI enoyl reductase from francisella tularensis: residence time and in vivo activity
- PMID: 19206187
- PMCID: PMC2693246
- DOI: 10.1021/cb800306y
Slow-onset inhibition of the FabI enoyl reductase from francisella tularensis: residence time and in vivo activity
Abstract
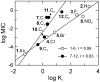
Francisella tularensis is a highly virulent and contagious Gram-negative intracellular bacterium that causes the disease tularemia in mammals. The high infectivity and the ability of the bacterium to survive for weeks in a cool, moist environment have raised the possibility that this organism could be exploited deliberately as a potential biological weapon. Fatty acid biosynthesis (FAS-II) is essential for bacterial viability and has been validated as a target for the discovery of novel antibacterials. The FAS-II enoyl reductase ftuFabI has been cloned and expressed, and a series of diphenyl ethers have been identified that are subnanomolar inhibitors of the enzyme with MIC90 values as low as 0.00018 microg mL(-1). The existence of a linear correlation between the Ki and MIC values strongly suggests that the antibacterial activity of the diphenyl ethers results from direct inhibition of ftuFabI within the cell. The compounds are slow-onset inhibitors of ftuFabI, and the residence time of the inhibitors on the enzyme correlates with their in vivo activity in a mouse model of tularemia infection. Significantly, the rate of breakdown of the enzyme-inhibitor complex is a better predictor of in vivo activity than the overall thermodynamic stability of the complex, a concept that has important implications for the discovery of novel chemotherapeutics that normally rely on equilibrium measurements of potency.
Figures







References
-
- Sjostedt A. Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect. 2006;8:561–567. - PubMed
-
- Oyston PC, Sjostedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol. 2004;2:967–978. - PubMed
-
- White SW, Zheng J, Zhang YM, Rock The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem. 2005;74:791–831. - PubMed
-
- Bergler H, Fuchsbichler S, Hogenauer G, Turnowsky F. The enoyl-[acyl-carrier-protein] reductase (FabI) of Escherichia coli, which catalyzes a key regulatory step in fatty acid biosynthesis, accepts NADH and NADPH as cofactors and is inhibited by palmitoyl-CoA. Eur J Biochem. 1996;242:689–694. - PubMed
-
- Zhang YM, White SW, Rock CO. Inhibiting bacterial fatty acid synthesis. J Biol Chem. 2006;281:17541–17544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Medical
Research Materials
Miscellaneous

