DNA recognition by the DNA primase of bacteriophage T7: a structure-function study of the zinc-binding domain
- PMID: 19206208
- PMCID: PMC2880633
- DOI: 10.1021/bi802123t
DNA recognition by the DNA primase of bacteriophage T7: a structure-function study of the zinc-binding domain
Abstract
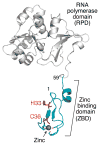
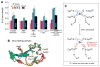
Synthesis of oligoribonucleotide primers for lagging-strand DNA synthesis in the DNA replication system of bacteriophage T7 is catalyzed by the primase domain of the gene 4 helicase-primase. The primase consists of a zinc-binding domain (ZBD) and an RNA polymerase (RPD) domain. The ZBD is responsible for recognition of a specific sequence in the ssDNA template whereas catalytic activity resides in the RPD. The ZBD contains a zinc ion coordinated with four cysteine residues. We have examined the ligation state of the zinc ion by X-ray absorption spectroscopy and biochemical analysis of genetically altered primases. The ZBD of primase engaged in catalysis exhibits considerable asymmetry in coordination to zinc, as evidenced by a gradual increase in electron density of the zinc together with elongation of the zinc-sulfur bonds. Both wild-type primase and primase reconstituted from purified ZBD and RPD have a similar electronic change in the level of the zinc ion as well as the configuration of the ZBD. Single amino acid replacements in the ZBD (H33A and C36S) result in the loss of both zinc binding and its structural integrity. Thus the zinc in the ZBD may act as a charge modulation indicator for the surrounding sulfur atoms necessary for recognition of specific DNA sequences.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

