Towards comprehensive structural motif mining for better fold annotation in the "twilight zone" of sequence dissimilarity
- PMID: 19208148
- PMCID: PMC2648771
- DOI: 10.1186/1471-2105-10-S1-S46
Towards comprehensive structural motif mining for better fold annotation in the "twilight zone" of sequence dissimilarity
Abstract
Background: Automatic identification of structure fingerprints from a group of diverse protein structures is challenging, especially for proteins whose divergent amino acid sequences may fall into the "twilight-" or "midnight-" zones where pair-wise sequence identities to known sequences fall below 25% and sequence-based functional annotations often fail.
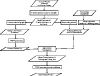
Results: Here we report a novel graph database mining method and demonstrate its application to protein structure pattern identification and structure classification. The biologic motivation of our study is to recognize common structure patterns in "immunoevasins", proteins mediating virus evasion of host immune defense. Our experimental study, using both viral and non-viral proteins, demonstrates the efficiency and efficacy of the proposed method.
Conclusion: We present a theoretic framework, offer a practical software implementation for incorporating prior domain knowledge, such as substitution matrices as studied here, and devise an efficient algorithm to identify approximate matched frequent subgraphs. By doing so, we significantly expanded the analytical power of sophisticated data mining algorithms in dealing with large volume of complicated and noisy protein structure data. And without loss of generality, choice of appropriate compatibility matrices allows our method to be easily employed in domains where subgraph labels have some uncertainty.
Figures
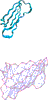
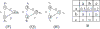

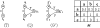

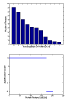
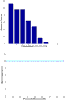
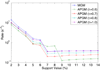
Similar articles
-
PSS-3D1D: an improved 3D1D profile method of protein fold recognition for the annotation of twilight zone sequences.J Struct Funct Genomics. 2011 Dec;12(4):181-9. doi: 10.1007/s10969-011-9119-x. Epub 2011 Dec 3. J Struct Funct Genomics. 2011. PMID: 22160493
-
Atomic interaction networks in the core of protein domains and their native folds.PLoS One. 2010 Feb 23;5(2):e9391. doi: 10.1371/journal.pone.0009391. PLoS One. 2010. PMID: 20186337 Free PMC article.
-
Structure motif discovery and mining the PDB.Bioinformatics. 2002 Feb;18(2):362-7. doi: 10.1093/bioinformatics/18.2.362. Bioinformatics. 2002. PMID: 11847094
-
General overview on structure prediction of twilight-zone proteins.Theor Biol Med Model. 2015 Sep 4;12:15. doi: 10.1186/s12976-015-0014-1. Theor Biol Med Model. 2015. PMID: 26338054 Free PMC article. Review.
-
Computational prediction of protein-protein interactions.Methods Mol Biol. 2004;261:445-68. doi: 10.1385/1-59259-762-9:445. Methods Mol Biol. 2004. PMID: 15064475 Review.
Cited by
-
PSS-3D1D: an improved 3D1D profile method of protein fold recognition for the annotation of twilight zone sequences.J Struct Funct Genomics. 2011 Dec;12(4):181-9. doi: 10.1007/s10969-011-9119-x. Epub 2011 Dec 3. J Struct Funct Genomics. 2011. PMID: 22160493
References
-
- RF D. Of URFs and ORFs: A Primer on How to Analyze Derived Amino Acid Sequences. Vol. 92. Mill Valley: University Science Books; 1986.
-
- B R. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. - PubMed
-
- JU B, R L, D E. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 253:164–170. 1991 Jul 12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

