Ontology-driven indexing of public datasets for translational bioinformatics
- PMID: 19208184
- PMCID: PMC2646250
- DOI: 10.1186/1471-2105-10-S2-S1
Ontology-driven indexing of public datasets for translational bioinformatics
Abstract
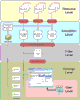
The volume of publicly available genomic scale data is increasing. Genomic datasets in public repositories are annotated with free-text fields describing the pathological state of the studied sample. These annotations are not mapped to concepts in any ontology, making it difficult to integrate these datasets across repositories. We have previously developed methods to map text-annotations of tissue microarrays to concepts in the NCI thesaurus and SNOMED-CT. In this work we generalize our methods to map text annotations of gene expression datasets to concepts in the UMLS. We demonstrate the utility of our methods by processing annotations of datasets in the Gene Expression Omnibus. We demonstrate that we enable ontology-based querying and integration of tissue and gene expression microarray data. We enable identification of datasets on specific diseases across both repositories. Our approach provides the basis for ontology-driven data integration for translational research on gene and protein expression data. Based on this work we have built a prototype system for ontology based annotation and indexing of biomedical data. The system processes the text metadata of diverse resource elements such as gene expression data sets, descriptions of radiology images, clinical-trial reports, and PubMed article abstracts to annotate and index them with concepts from appropriate ontologies. The key functionality of this system is to enable users to locate biomedical data resources related to particular ontology concepts.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

