EpiGRAPH: user-friendly software for statistical analysis and prediction of (epi)genomic data
- PMID: 19208250
- PMCID: PMC2688269
- DOI: 10.1186/gb-2009-10-2-r14
EpiGRAPH: user-friendly software for statistical analysis and prediction of (epi)genomic data
Abstract
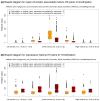
The EpiGRAPH web service http://epigraph.mpi-inf.mpg.de/ enables biologists to uncover hidden associations in vertebrate genome and epigenome datasets. Users can upload sets of genomic regions and EpiGRAPH will test multiple attributes (including DNA sequence, chromatin structure, epigenetic modifications and evolutionary conservation) for enrichment or depletion among these regions. Furthermore, EpiGRAPH learns to predictively identify similar genomic regions. This paper demonstrates EpiGRAPH's practical utility in a case study on monoallelic gene expression and describes its novel approach to reproducible bioinformatic analysis.
Figures




References
-
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

