Inducible expression of beta defensins by human respiratory epithelial cells exposed to Aspergillus fumigatus organisms
- PMID: 19208266
- PMCID: PMC2653505
- DOI: 10.1186/1471-2180-9-33
Inducible expression of beta defensins by human respiratory epithelial cells exposed to Aspergillus fumigatus organisms
Abstract
Background: Aspergillus fumigatus, a saprophytic mould, is responsible for life-threatening, invasive pulmonary diseases in immunocompromised hosts. The role of the airway epithelium involves a complex interaction with the inhaled pathogen. Antimicrobial peptides with direct antifungal and chemotactic activities may boost antifungal immune response.
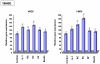
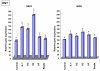
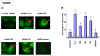
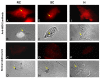
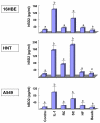
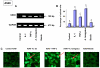
Results: The inducible expression of defensins by human bronchial epithelial 16HBE cells and A549 pneumocyte cells exposed to A. fumigatus was investigated. Using RT-PCR and real time PCR, we showed an activation of hBD2 and hBD9 defensin genes: the expression was higher in cells exposed to swollen conidia (SC), compared to resting conidia (RC) or hyphal fragments (HF). The kinetics of defensin expression was different for each one, evoking a putative distinct function for each investigated defensin. The decrease of defensin expression in the presence of heat-inactivated serum indicated a possible link between defensins and the proteins of the host complement system. The presence of defensin peptide hBD2 was revealed using immunofluorescence that showed a punctual cytoplasmic and perinuclear staining. Quantification of the cells stained with anti hBD2 antibody demonstrated that SC induced a greater number of cells that synthesized hBD2, compared to RC or HF. Labelling of the cells with anti-hBD-2 antibody showed a positive immunofluorescence signal around RC or SC in contrast to HF. This suggests co-localisation of hBD2 and digested conidia. The HBD2 level was highest in the supernatants of cells exposed to SC, as was determined by sandwich ELISA. Experiments using neutralising anti-interleukine-1beta antibody reflect the autocrine mechanism of defensin expression induced by SC. Investigation of defensin expression at transcriptional and post-transcriptional levels demonstrated the requirement of transcription as well as new protein synthesis during A. fumigatus defensin induction. Finally, induced defensin expression in primary culture of human respiratory cells exposed to A. fumigatus points to the biological significance of described phenomena.
Conclusion: Our findings provide evidence that respiratory epithelium might play an important role in the immune response during Aspergillus infection. Understanding the mechanisms of regulation of defensin expression may thus lead to new approaches that could enhance expression of antimicrobial peptides for potential therapeutic use during aspergillosis treatment.
Figures






References
-
- Ganz T, Weiss T. Antimicrobial peptides of phagocytes and epithelia. Semin Hematol. 1997;34(4):343–354. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

