Pax6 regulation of Math5 during mouse retinal neurogenesis
- PMID: 19208436
- PMCID: PMC2976517
- DOI: 10.1002/dvg.20479
Pax6 regulation of Math5 during mouse retinal neurogenesis
Abstract
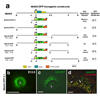
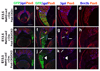
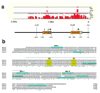
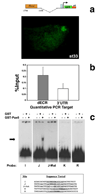
Activation of the bHLH factor Math5 (Atoh7) is an initiating event for mammalian retinal neurogenesis, as it is critically required for retinal ganglion cell formation. However, the cis-regulatory elements and trans-acting factors that control Math5 expression are largely unknown. Using a combination of transgenic mice and bioinformatics, we identified a phylogenetically conserved regulatory element that is required to activate Math5 transcription during early retinal neurogenesis. This element drives retinal expression in vivo, in a cross-species transgenic assay. Previously, Pax6 was shown to be necessary for the initiation of Math5 mRNA expression. We extend this finding by showing that the Math5 retinal enhancer also requires Pax6 for its activation, via Pax6 binding to a highly conserved binding site. In addition, our data reveal that other retinal factors are required for accurate regulation of Math5 by Pax6.
Figures

Similar articles
-
Conserved regulation of Math5 and Math1 revealed by Math5-GFP transgenes.Mol Cell Neurosci. 2007 Dec;36(4):435-48. doi: 10.1016/j.mcn.2007.08.006. Epub 2007 Aug 15. Mol Cell Neurosci. 2007. PMID: 17900924 Free PMC article.
-
Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis.Development. 1998 Dec;125(23):4821-33. doi: 10.1242/dev.125.23.4821. Development. 1998. PMID: 9806930
-
Multiple requirements for Hes 1 during early eye formation.Dev Biol. 2005 Aug 15;284(2):464-78. doi: 10.1016/j.ydbio.2005.06.010. Dev Biol. 2005. PMID: 16038893 Free PMC article.
-
Role of Fabp7, a downstream gene of Pax6, in the maintenance of neuroepithelial cells during early embryonic development of the rat cortex.J Neurosci. 2005 Oct 19;25(42):9752-61. doi: 10.1523/JNEUROSCI.2512-05.2005. J Neurosci. 2005. PMID: 16237179 Free PMC article.
-
Developmental malformations of the eye: the role of PAX6, SOX2 and OTX2.Clin Genet. 2006 Jun;69(6):459-70. doi: 10.1111/j.1399-0004.2006.00619.x. Clin Genet. 2006. PMID: 16712695 Review.
Cited by
-
Cloning and Functional Analysis of Pax6 from the Hydrothermal Vent Tubeworm Ridgeia piscesae.PLoS One. 2016 Dec 22;11(12):e0168579. doi: 10.1371/journal.pone.0168579. eCollection 2016. PLoS One. 2016. PMID: 28005979 Free PMC article.
-
Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease.Nat Neurosci. 2011 May;14(5):578-86. doi: 10.1038/nn.2798. Epub 2011 Mar 27. Nat Neurosci. 2011. PMID: 21441919 Free PMC article.
-
Integrated copy number and gene expression analysis detects a CREB1 association with Alzheimer's disease.Transl Psychiatry. 2012 Nov 20;2(11):e192. doi: 10.1038/tp.2012.119. Transl Psychiatry. 2012. PMID: 23168992 Free PMC article.
-
Heterochronic misexpression of Ascl1 in the Atoh7 retinal cell lineage blocks cell cycle exit.Mol Cell Neurosci. 2013 May;54:108-20. doi: 10.1016/j.mcn.2013.02.004. Epub 2013 Feb 26. Mol Cell Neurosci. 2013. PMID: 23481413 Free PMC article.
-
Key transcription factors influence the epigenetic landscape to regulate retinal cell differentiation.Nucleic Acids Res. 2023 Mar 21;51(5):2151-2176. doi: 10.1093/nar/gkad026. Nucleic Acids Res. 2023. PMID: 36715342 Free PMC article.
References
-
- Amaya E, Kroll KL. A method for generating transgenic frog embryos. Methods Mol Biol. 1999;97:393–414. - PubMed
-
- Austin CP, Feldman DE, Ida JA, Jr, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. - PubMed
-
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. - PubMed
-
- Brzezinski JA. Department of Human Genetics. Ann Arbor, MI: University of Michigan; 2005. The Role of Math5 in Retinal Development.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

