MitoMiner, an integrated database for the storage and analysis of mitochondrial proteomics data
- PMID: 19208617
- PMCID: PMC2690483
- DOI: 10.1074/mcp.M800373-MCP200
MitoMiner, an integrated database for the storage and analysis of mitochondrial proteomics data
Abstract
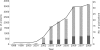
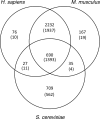
Mitochondria are a vital component of eukaryotic cells with functions that extend beyond energy production to include metabolism, signaling, cell growth, and apoptosis. Their dysfunction is implicated in a large number of metabolic, degenerative, and age-related human diseases. Therefore, it is important to characterize and understand the mitochondrion. Many experiments have attempted to define the mitochondrial proteome, resulting in large and complex data sets that are difficult to analyze. To address this, we developed a new public resource for the storage and investigation of this mitochondrial proteomics data, called MitoMiner, that uses a model to describe the proteomics data and associated biological information. The proteomics data of 33 publications from both mass spectrometry and green fluorescent protein tagging experiments were imported and integrated with protein annotation from UniProt and genome projects, metabolic pathway data from Kyoto Encyclopedia of Genes and Genomes, homology relationships from HomoloGene, and disease information from Online Mendelian Inheritance in Man. We demonstrate the strengths of MitoMiner by investigating these data sets and show that the number of different mitochondrial proteins that have been reported is about 3700, although the number of proteins common to both animals and yeast is about 1400, and membrane proteins appear to be underrepresented. Furthermore analysis indicated that enzymes of some cytosolic metabolic pathways are regularly detected in mitochondrial proteomics experiments, suggesting that they are associated with the outside of the outer mitochondrial membrane. The data and advanced capabilities of MitoMiner provide a framework for further mitochondrial analysis and future systems level modeling of mitochondrial physiology.
Figures







Similar articles
-
MitoMiner: a data warehouse for mitochondrial proteomics data.Nucleic Acids Res. 2012 Jan;40(Database issue):D1160-7. doi: 10.1093/nar/gkr1101. Epub 2011 Nov 24. Nucleic Acids Res. 2012. PMID: 22121219 Free PMC article.
-
MitoMiner v3.1, an update on the mitochondrial proteomics database.Nucleic Acids Res. 2016 Jan 4;44(D1):D1258-61. doi: 10.1093/nar/gkv1001. Epub 2015 Oct 1. Nucleic Acids Res. 2016. PMID: 26432830 Free PMC article.
-
MitoMiner v4.0: an updated database of mitochondrial localization evidence, phenotypes and diseases.Nucleic Acids Res. 2019 Jan 8;47(D1):D1225-D1228. doi: 10.1093/nar/gky1072. Nucleic Acids Res. 2019. PMID: 30398659 Free PMC article.
-
Mitochondrial proteomics--a tool for the study of metabolic disorders.J Inherit Metab Dis. 2012 Jul;35(4):715-26. doi: 10.1007/s10545-012-9480-3. Epub 2012 Apr 17. J Inherit Metab Dis. 2012. PMID: 22526845 Review.
-
Mammalian mitochondrial proteomics: insights into mitochondrial functions and mitochondria-related diseases.Expert Rev Proteomics. 2010 Jun;7(3):333-45. doi: 10.1586/epr.10.22. Expert Rev Proteomics. 2010. PMID: 20536306 Review.
Cited by
-
Perspectives on: SGP symposium on mitochondrial physiology and medicine: mitochondrial proteome design: from molecular identity to pathophysiological regulation.J Gen Physiol. 2012 Jun;139(6):395-406. doi: 10.1085/jgp.201210797. J Gen Physiol. 2012. PMID: 22641634 Free PMC article. No abstract available.
-
Guidelines for mitochondrial RNA analysis.Mol Ther Nucleic Acids. 2024 Jun 26;35(3):102262. doi: 10.1016/j.omtn.2024.102262. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39091381 Free PMC article. Review.
-
Is ABCC6 a genuine mitochondrial protein?BMC Res Notes. 2013 Oct 23;6:427. doi: 10.1186/1756-0500-6-427. BMC Res Notes. 2013. PMID: 24152371 Free PMC article.
-
Visualization and biochemical analyses of the emerging mammalian 14-3-3-phosphoproteome.Mol Cell Proteomics. 2011 Oct;10(10):M110.005751. doi: 10.1074/mcp.M110.005751. Epub 2011 Jul 1. Mol Cell Proteomics. 2011. PMID: 21725060 Free PMC article.
-
Proteomics as a Tool for the Study of Mitochondrial Proteome, Its Dysfunctionality and Pathological Consequences in Cardiovascular Diseases.Int J Mol Sci. 2023 Feb 28;24(5):4692. doi: 10.3390/ijms24054692. Int J Mol Sci. 2023. PMID: 36902123 Free PMC article. Review.
References
-
- DiMauro, S., and Schon, E. A. ( 2003) Mitochondrial respiratory-chain diseases. N. Eng. J. Med. 348, 2656–2668 - PubMed
-
- Trifunovic, A., Wredenberg, A., Falkenberg, M., Spelbrink, J. N., Rovio, A. T., Bruder, C. E., Bohlooly, Y. M., Gidlof, S., Oldfors, A., Wibom, R., Tornell, J., Jacobs, H. T., and Larsson, N. G. ( 2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 - PubMed
-
- Fliss, M. S., Usadel, H., Caballero, O. L., Wu, L., Buta, M. R., Eleff, S. M., Jen, J., and Sidransky, D. ( 2000) Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science 287, 2017–2019 - PubMed
-
- Taylor, S. W., Fahy, E., and Ghosh, S. S. ( 2003) Global organellar proteomics. Trends Biotechnol. 21, 82–88 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

