Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen
- PMID: 19208623
- PMCID: PMC2667742
- DOI: 10.1074/jbc.M809745200
Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen
Abstract
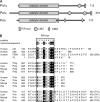
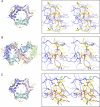
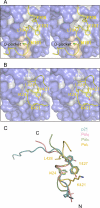
Translesion synthesis (TLS) is a DNA damage tolerance mechanism that allows continued DNA synthesis, even in the presence of damaged DNA templates. Mammals have multiple DNA polymerases specialized for TLS, including Poleta, Poliota, and Polkappa. These enzymes show preferential bypass for different lesions. Proliferating cell nuclear antigen (PCNA), which functions as a sliding clamp for the replicative polymerase Poldelta, also interacts with the three TLS polymerases. Although many PCNA-binding proteins have a highly conserved sequence termed the PCNA-interacting protein box (PIP-box), Poleta, Poliota, and Polkappa have a noncanonical PIP-box sequence. In response to DNA damage, Lys-164 of PCNA undergoes ubiquitination by the RAD6-RAD18 complex, and the ubiquitination is considered to facilitate TLS. Consistent with this, these three TLS polymerases have one or two ubiquitin binding domains and are recruited to replication forks via interactions with ubiquitinated PCNA involving the noncanonical PIP-box and ubiquitin binding domain. However, it is unclear how these TLS polymerases interact with PCNA. To address the structural basis for interactions between different TLS polymerases and PCNA, we determined crystal structures of PCNA bound to peptides containing the noncanonical PIP-box of these polymerases. We show that the three PIP-box peptides interact with PCNA in different ways, both from one another and from canonical PIP-box peptides. Especially, the PIP-box of Poliota adopts a novel structure. Furthermore, these structures enable us to speculate how these TLS polymerases interact with Lys-164-monoubiquitinated PCNA. Our results will provide clues to understanding the mechanism of preferential recruitment of TLS polymerases to the stalled forks.
Figures
Similar articles
-
NMR mapping of PCNA interaction with translesion synthesis DNA polymerase Rev1 mediated by Rev1-BRCT domain.J Mol Biol. 2013 Sep 9;425(17):3091-105. doi: 10.1016/j.jmb.2013.05.029. Epub 2013 Jun 7. J Mol Biol. 2013. PMID: 23747975
-
Initial crystallographic study of human PCNA in complex with a peptide containing the noncanonical PIP-box sequence of human DNA polymerase iota.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008 Oct 1;64(Pt 10):954-6. doi: 10.1107/S1744309108028522. Epub 2008 Sep 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008. PMID: 18931444 Free PMC article.
-
Different types of interaction between PCNA and PIP boxes contribute to distinct cellular functions of Y-family DNA polymerases.Nucleic Acids Res. 2015 Sep 18;43(16):7898-910. doi: 10.1093/nar/gkv712. Epub 2015 Jul 13. Nucleic Acids Res. 2015. PMID: 26170230 Free PMC article.
-
Ubiquitin-dependent regulation of translesion polymerases.Biochem Soc Trans. 2010 Feb;38(Pt 1):110-5. doi: 10.1042/BST0380110. Biochem Soc Trans. 2010. PMID: 20074045 Review.
-
Regulatory role of ubiquitin in eukaryotic DNA translesion synthesis.Biochemistry. 2013 May 14;52(19):3217-28. doi: 10.1021/bi400194r. Epub 2013 May 1. Biochemistry. 2013. PMID: 23634825 Review.
Cited by
-
A non-catalytic role of DNA polymerase η in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks.Nucleic Acids Res. 2013 Mar 1;41(5):3079-93. doi: 10.1093/nar/gkt016. Epub 2013 Jan 23. Nucleic Acids Res. 2013. PMID: 23345618 Free PMC article.
-
Replication-independent repair of DNA interstrand crosslinks.Mol Cell. 2012 Jul 13;47(1):140-7. doi: 10.1016/j.molcel.2012.05.001. Epub 2012 May 31. Mol Cell. 2012. PMID: 22658724 Free PMC article.
-
Role of DNA polymerase κ in the maintenance of genomic stability.Mol Cell Oncol. 2014 Jul 15;1(1):e29902. doi: 10.4161/mco.29902. eCollection 2014. Mol Cell Oncol. 2014. PMID: 27308312 Free PMC article. Review.
-
ELM-the eukaryotic linear motif resource in 2020.Nucleic Acids Res. 2020 Jan 8;48(D1):D296-D306. doi: 10.1093/nar/gkz1030. Nucleic Acids Res. 2020. PMID: 31680160 Free PMC article.
-
Cryo-EM structure of human Pol κ bound to DNA and mono-ubiquitylated PCNA.Nat Commun. 2021 Oct 19;12(1):6095. doi: 10.1038/s41467-021-26251-6. Nat Commun. 2021. PMID: 34667155 Free PMC article.
References
-
- Ohmori, H., Friedberg, E. C., Fuchs, R. P., Goodman, M. F., Hanaoka, F., Hinkle, D., Kunkel, T. A., Lawrence, C. W., Livneh, Z., Nohmi, T., Prakash, L., Prakash, S., Todo, T., Walker, G. C., Wang, Z., and Woodgate, R. (2001) Mol. Cell 8 7-8 - PubMed
-
- Vaisman, A., Lehmann, A. R., and Woodgate, R. (2004) Adv. Protein Chem. 69 205-228 - PubMed
-
- Ohmori, H., Ohashi, E., and Ogi, T. (2004) Adv. Protein Chem. 69 265-278 - PubMed
-
- Masutani, C., Kusumoto, R., Yamada, A., Dohmae, N., Yokoi, M., Yuasa, M., Araki, M., Iwai, S., Takio, K., and Hanaoka, F. (1999) Nature 399 700-704 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

