Snake venom Vascular Endothelial Growth Factors (VEGF-Fs) exclusively vary their structures and functions among species
- PMID: 19208624
- PMCID: PMC2665111
- DOI: 10.1074/jbc.M809071200
Snake venom Vascular Endothelial Growth Factors (VEGF-Fs) exclusively vary their structures and functions among species
Abstract
Vascular endothelial growth factor (VEGF-A) and its family proteins are crucial regulators of blood vessel formation and vascular permeability. Snake venom has recently been shown to be an exogenous source of unique VEGF (known as VEGF-F), and now, two types of VEGF-F with distinct biochemical properties have been reported. Here, we show that VEGF-Fs (venom-type VEGFs) are highly variable in structure and function among species, in contrast to endogenous tissue-type VEGFs (VEGF-As) of snakes. Although the structures of tissue-type VEGFs are highly conserved among venomous snake species and even among all vertebrates, including humans, those of venom-type VEGFs are extensively variegated, especially in the regions around receptor-binding loops and C-terminal putative coreceptor-binding regions, indicating that highly frequent variations are located around functionally key regions of the proteins. Genetic analyses suggest that venom-type VEGF gene may have developed from a tissue-type gene and that the unique sequence of its C-terminal region was generated by an alteration in the translation frame in the corresponding exons. We further verified that a novel venom-type VEGF from Bitis arietans displays unique properties distinct from already known VEGFs. Our results may provide evidence of a novel mechanism causing the generation of multiple snake toxins and also of a new model of molecular evolution.
Figures
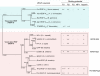
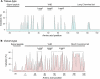
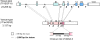


References
-
- Olsson, A. K., Dimberg, A., Kreuger, J., and Claesson-Welsh, L. (2006) Nat. Rev. Mol. Cell Biol. 7 359–371 - PubMed
-
- Yamazaki, Y., and Morita, T. (2006) Mol. Divers. 10 515–527 - PubMed
-
- Carmeliet, P. (2003) Nat. Rev. Genet. 4 710–720 - PubMed
-
- Yamazaki, Y., and Morita, T. (2007) Curr. Pharm. Des. 13 2872–2886 - PubMed
-
- Yamazaki, Y., Takani, K., Atoda, H., and Morita, T. (2003) J. Biol. Chem. 278 51985–51988 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

