Stimulation of UvrD helicase by UvrAB
- PMID: 19208629
- PMCID: PMC2666613
- DOI: 10.1074/jbc.M808030200
Stimulation of UvrD helicase by UvrAB
Abstract
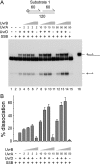
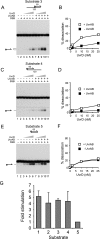
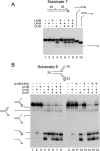
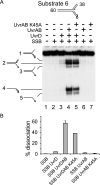
Helicases play critical roles in all aspects of nucleic acid metabolism by catalyzing the remodeling of DNA and RNA structures. UvrD is an abundant helicase in Escherichia coli with well characterized functions in mismatch and nucleotide excision repair and a possible role in displacement of proteins such as RecA from single-stranded DNA. The mismatch repair protein MutL is known to stimulate UvrD. Here we show that the nucleotide excision repair proteins UvrA and UvrB can together stimulate UvrD-catalyzed unwinding of a range of DNA substrates containing strand discontinuities, including forked DNA substrates. The stimulation is specific for UvrD, as UvrAB failed to stimulate Rep helicase, a UvrD homologue. Moreover, although UvrAB can promote limited strand displacement, stimulation of UvrD did not require the strand displacement function of UvrAB. We conclude that UvrAB, like MutL, modulate UvrD helicase activity. This stimulation likely plays a role in DNA strand and protein displacement by UvrD in nucleotide excision repair. Promotion of UvrD-catalyzed unwinding of nicked duplexes by UvrAB may also explain the need for UvrAB and UvrD in Okazaki fragment processing in cells lacking DNA polymerase I. More generally, these data support the idea that helicase activity is regulated in vivo, with helicases acting as part of multisubunit complexes rather than in isolation.
Figures




References
-
- Subramanya, H. S., Bird, L. E., Brannigan, J. A., and Wigley, D. B. (1996) Nature 384 379-383 - PubMed
-
- Korolev, S., Hsieh, J., Gauss, G. H., Lohman, T. M., and Waksman, G. (1997) Cell 90 635-647 - PubMed
-
- Singleton, M. R., Dillingham, M. S., and Wigley, D. B. (2007) Annu. Rev. Biochem. 76 23-50 - PubMed
-
- Wahle, E., Lasken, R. S., and Kornberg, A. (1989) J. Biol. Chem. 264 2469-2475 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

