Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms
- PMID: 19208647
- PMCID: PMC2647321
- DOI: 10.1093/nar/gkp032
Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms
Abstract
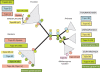
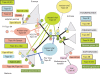
Topoisomerases are essential enzymes that solve topological problems arising from the double-helical structure of DNA. As a consequence, one should have naively expected to find homologous topoisomerases in all cellular organisms, dating back to their last common ancestor. However, as observed for other enzymes working with DNA, this is not the case. Phylogenomics analyses indicate that different sets of topoisomerases were present in the most recent common ancestors of each of the three cellular domains of life (some of them being common to two or three domains), whereas other topoisomerases families or subfamilies were acquired in a particular domain, or even a particular lineage, by horizontal gene transfers. Interestingly, two groups of viruses encode topoisomerases that are only distantly related to their cellular counterparts. To explain these observations, we suggest that topoisomerases originated in an ancestral virosphere, and that various subfamilies were later on transferred independently to different ancient cellular lineages. We also proposed that topoisomerases have played a critical role in the origin of modern genomes and in the emergence of the three cellular domains.
Figures


References
-
- Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. - PubMed
-
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. - PubMed
-
- Lazcano A, Guerrero R, Margulis L, Oró J. The evolutionary transition from RNA to DNA in early cells. J. Mol. Evol. 1988;27:283–290. - PubMed
-
- Gadelle D, Filée J, Buhler C, Forterre P. Phylogenomics of type II DNA topoisomerases. Bioessays. 2003;3:232–242. - PubMed
-
- Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;4:427–446. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

