Inactivation of the MAL gene in breast cancer is a common event that predicts benefit from adjuvant chemotherapy
- PMID: 19208741
- PMCID: PMC2700346
- DOI: 10.1158/1541-7786.MCR-08-0314
Inactivation of the MAL gene in breast cancer is a common event that predicts benefit from adjuvant chemotherapy
Abstract
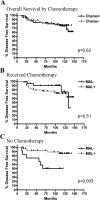
Dysregulation of MAL (myelin and lymphocyte protein) has been implicated in several malignancies including esophageal, ovarian, and cervical cancers. The MAL protein functions in apical transport in polarized epithelial cells; therefore, its disruption may lead to loss of organized polarity characteristic of most solid malignancies. Bisulfite sequencing of the MAL promoter CpG island revealed hypermethylation in breast cancer cell lines and 69% of primary tumors analyzed compared with normal breast epithelial cells. Differential methylation between normal and cancer DNA was confined to the proximal promoter region. In a subset of breast cancer cell lines including T47D and MCF7 cells, promoter methylation correlated with transcriptional silencing that was reversible with the methylation inhibitor 5-aza-2'-deoxycytidine. In addition, expression of MAL reduced motility and resulted in a redistribution of lipid raft components in MCF10A cells. MAL protein expression measured by immunohistochemistry revealed no significant correlation with clinicopathologic features. However, in patients who did not receive adjuvant chemotherapy, reduced MAL expression was a significant predictive factor for disease-free survival. These data implicate MAL as a commonly altered gene in breast cancer with implications for response to chemotherapy.
Figures




References
-
- Santini V, Kantarjian HM, Issa JP. Changes in DNA methylation in neoplasia: pathophysiology and therapeutic implications. Annals of internal medicine. 2001;134:573–86. - PubMed
-
- Szyf M, Pakneshan P, Rabbani SA. DNA demethylation and cancer: therapeutic implications. Cancer letters. 2004;211:133–43. - PubMed
-
- Dulaimi E, Hillinck J, Ibanez de Caceres I, Al-Saleem T, Cairns P. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin Cancer Res. 2004;10:6189–93. - PubMed
-
- Momparler RL. Cancer epigenetics. Oncogene. 2003;22:6479–83. - PubMed
-
- Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. The Journal of biological chemistry. 1997;272:22322–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

