Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression
- PMID: 19208811
- PMCID: PMC2638738
- DOI: 10.1073/pnas.0813345106
Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression
Abstract
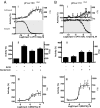
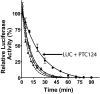
High-throughput screening (HTS) assays used in drug discovery frequently use reporter enzymes such as firefly luciferase (FLuc) as indicators of target activity. An important caveat to consider, however, is that compounds can directly affect the reporter, leading to nonspecific but highly reproducible assay signal modulation. In rare cases, this activity appears counterintuitive; for example, some FLuc inhibitors, acting through posttranslational Fluc reporter stabilization, appear to activate gene expression. Previous efforts to characterize molecules that influence luciferase activity identified a subset of 3,5-diaryl-oxadiazole-containing compounds as FLuc inhibitors. Here, we evaluate a number of compounds with this structural motif for activity against FLuc. One such compound is PTC124 {3-[5-(2-fluorophenyl)-1,2,4-oxadiazol-3-yl]benzoic acid}, a molecule originally identified in a cell-based FLuc assay as having nonsense codon suppression activity [Welch EM, et al., Nature (2007) 447:87-91]. We find that the potency of FLuc inhibition for the tested compounds strictly correlates with their activity in a FLuc reporter cell-based nonsense codon assay, with PTC124 emerging as the most potent FLuc inhibitor (IC(50) = 7 +/- 1 nM). However, these compounds, including PTC124, fail to show nonsense codon suppression activity when Renilla reniformis luciferase (RLuc) is used as a reporter and are inactive against the RLuc enzyme. This suggests that the initial discovery of PTC124 may have been biased by its direct effect on the FLuc reporter, implicating firefly luciferase as a molecular target of PTC124. Our results demonstrate the value of understanding potential interactions between reporter enzymes and chemical compounds and emphasize the importance of implementing the appropriate control assays before interpreting HTS results.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
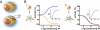

Comment in
-
Nonsense suppression activity of PTC124 (ataluren).Proc Natl Acad Sci U S A. 2009 Jun 23;106(25):E64; author reply E65. doi: 10.1073/pnas.0901936106. Epub 2009 Jun 8. Proc Natl Acad Sci U S A. 2009. PMID: 19506240 Free PMC article. No abstract available.
References
-
- Bender A, et al. Which aspects of HTS are empirically correlated with downstream success? Curr Opin Drug Discov Devel. 2008;11:327–337. - PubMed
-
- Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol. 2007;5:127–136. - PubMed
-
- Inglese J, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. - PubMed
-
- Thompson PA, et al. Identification of ligand binding by protein stabilization: Comparison of ATLAS with biophysical and enzymatic methods. Assay Drug Dev Technol. 2008;6:69–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

