What have we learned from the studies of two-state folders, and what are the unanswered questions about two-state protein folding?
- PMID: 19208936
- PMCID: PMC2822238
- DOI: 10.1088/1478-3975/6/1/015001
What have we learned from the studies of two-state folders, and what are the unanswered questions about two-state protein folding?
Abstract
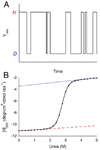
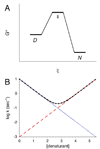
Small proteins with globular structures often fold by simple all-or-none mechanisms, both in an equilibrium and a kinetic sense, despite the very large number of partly folded conformations available. This type of 'two-state' folding will be discussed in terms of experimental tests, underlying molecular mechanisms, and limits to two-state behavior. Factors that appear to be important for two-state folding include topology (sequence distance of contacts in the native structure), molecular cooperativity and local energy distribution. Because their local stability distributions and cooperativities can be dissected and analyzed separately from topological features, recent studies of the folding of symmetric proteins will be discussed as a means to better understand the origins of two-state folding.
Figures
References
-
- Alm E, Morozov AV, Kortemme T, Baker D. Simple physical models connect theory and experiment in protein folding kinetics. J Mol Biol. 2002;322(2):463–476. - PubMed
-
- Anil B, Sato S, Cho JH, Raleigh DP. Fine structure analysis of a protein folding transition state; distinguishing between hydrophobic stabilization and specific packing. J Mol Biol. 2005;354(3):693–705. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
