The comprehensive native interactome of a fully functional tagged prion protein
- PMID: 19209230
- PMCID: PMC2635968
- DOI: 10.1371/journal.pone.0004446
The comprehensive native interactome of a fully functional tagged prion protein
Abstract
The enumeration of the interaction partners of the cellular prion protein, PrP(C), may help clarifying its elusive molecular function. Here we added a carboxy proximal myc epitope tag to PrP(C). When expressed in transgenic mice, PrP(myc) carried a GPI anchor, was targeted to lipid rafts, and was glycosylated similarly to PrP(C). PrP(myc) antagonized the toxicity of truncated PrP, restored prion infectibility of PrP(C)-deficient mice, and was physically incorporated into PrP(Sc) aggregates, indicating that it possessed all functional characteristics of genuine PrP(C). We then immunopurified myc epitope-containing protein complexes from PrP(myc) transgenic mouse brains. Gentle differential elution with epitope-mimetic decapeptides, or a scrambled version thereof, yielded 96 specifically released proteins. Quantitative mass spectrometry with isotope-coded tags identified seven proteins which co-eluted equimolarly with PrP(C) and may represent component of a multiprotein complex. Selected PrP(C) interactors were validated using independent methods. Several of these proteins appear to exert functions in axomyelinic maintenance.
Conflict of interest statement
Figures
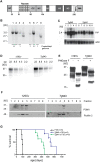
 mouse co-expressing N-proximally truncated PrPΔF. Lane 8: PrPΔF mouse. The bands diagnostic for PrPmyc and PrPΔF were 3039 and 2709 bp, respectively. Numbers of transgenic copies per haploid genome, as determined by quantitation of Southern blot signals against the respective Prnp
o genomic band, revealed higher copy numbers in Tg940 PrPmyc (#6) than in Tg941 PrPmyc mice (#1). (C) Northern blot analysis of individual Tg940 PrPmyc and Tg941 PrPmyc brains using a Prnp probe. Mice homozygous for the transgenic allele PrPmyc (lanes 2, 3, 4 from Tg940 and lanes 8, 11 from Tg941) showed higher levels of PrPmyc mRNA than hemizygous mice (lanes 1 and 5 from Tg940 and lanes 6, 9, 10 from Tg941). An actin probe was used as a loading control (lower panel). (D) Similar expression levels of transgenic protein from Tg940
mouse co-expressing N-proximally truncated PrPΔF. Lane 8: PrPΔF mouse. The bands diagnostic for PrPmyc and PrPΔF were 3039 and 2709 bp, respectively. Numbers of transgenic copies per haploid genome, as determined by quantitation of Southern blot signals against the respective Prnp
o genomic band, revealed higher copy numbers in Tg940 PrPmyc (#6) than in Tg941 PrPmyc mice (#1). (C) Northern blot analysis of individual Tg940 PrPmyc and Tg941 PrPmyc brains using a Prnp probe. Mice homozygous for the transgenic allele PrPmyc (lanes 2, 3, 4 from Tg940 and lanes 8, 11 from Tg941) showed higher levels of PrPmyc mRNA than hemizygous mice (lanes 1 and 5 from Tg940 and lanes 6, 9, 10 from Tg941). An actin probe was used as a loading control (lower panel). (D) Similar expression levels of transgenic protein from Tg940  , and full-length PrP from 129S2/SvPas wild-type mice, analyzed by Western blotting of total brain homogenate using anti-PrP antibody POM1. (E) Similar glycosylation pattern of full-length PrP from 129S2/SvPas wild-type and PrPmyc from Tg940
, and full-length PrP from 129S2/SvPas wild-type mice, analyzed by Western blotting of total brain homogenate using anti-PrP antibody POM1. (E) Similar glycosylation pattern of full-length PrP from 129S2/SvPas wild-type and PrPmyc from Tg940  mice. Brain homogenates were subjected to PNGase F treatment as indicated, and analyzed by Western blotting using POM1 antibody to PrPC. (F) Detergent-resistant membrane preparations from cerebella of Tg940 PrPmyc transgenic mice showed PrPmyc in lipid rafts. PrPmyc was detectable by Western blotting in fractions with 5–30% Optiprep. PrPmyc resided in the same fractions as flotillin (48 kDa) confirming its localization in DRMs. (G) A genetic in vivo assay for the function of the PrPmyc protein. Survival curves of mice expressing PrPΔF in absence of full length PrPC and in presence of PrPmyc from two transgenic lines. Toxicity of PrPΔF was counteracted by PrPmyc, leading to a longer survival and suggesting that PrPmyc has retained at least some of the function of PrPC. Line PrPΔF, Tg940 and Tg941 consisted of 5, 5, and 9 individuals, respectively.
mice. Brain homogenates were subjected to PNGase F treatment as indicated, and analyzed by Western blotting using POM1 antibody to PrPC. (F) Detergent-resistant membrane preparations from cerebella of Tg940 PrPmyc transgenic mice showed PrPmyc in lipid rafts. PrPmyc was detectable by Western blotting in fractions with 5–30% Optiprep. PrPmyc resided in the same fractions as flotillin (48 kDa) confirming its localization in DRMs. (G) A genetic in vivo assay for the function of the PrPmyc protein. Survival curves of mice expressing PrPΔF in absence of full length PrPC and in presence of PrPmyc from two transgenic lines. Toxicity of PrPΔF was counteracted by PrPmyc, leading to a longer survival and suggesting that PrPmyc has retained at least some of the function of PrPC. Line PrPΔF, Tg940 and Tg941 consisted of 5, 5, and 9 individuals, respectively.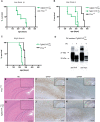
 mice and Prnp
+/o mice low dose ip inoculated with RML5 prions. Groups Tg940
mice and Prnp
+/o mice low dose ip inoculated with RML5 prions. Groups Tg940  ip Prnp
+/o ip consisted of 5 and 2 individuals, respectively. (B) Survival curves of Tg940
ip Prnp
+/o ip consisted of 5 and 2 individuals, respectively. (B) Survival curves of Tg940  , Tg941
, Tg941  , and Prnp
+/o mice inoculated low dose ic with RML5 prions. Group Tg940
, and Prnp
+/o mice inoculated low dose ic with RML5 prions. Group Tg940  comprises 6, group Tg941
comprises 6, group Tg941  4, and Prnp
+/o 3 individuals, respectively. (C) Survival curves of Tg940
4, and Prnp
+/o 3 individuals, respectively. (C) Survival curves of Tg940  mice and Prnp
+/o mice high dose ic inoculated with RML5 prions. Line Tg940
mice and Prnp
+/o mice high dose ic inoculated with RML5 prions. Line Tg940  comprises of 8 and line Prnp
+/o of 6 individuals, respectively. (D) PrPmyc was converted into myc-tagged proteinase K-resistant
comprises of 8 and line Prnp
+/o of 6 individuals, respectively. (D) PrPmyc was converted into myc-tagged proteinase K-resistant  in presence of a wild-type PrP allele. Western blot analysis using brain homogenate from an inoculated, terminally sick
in presence of a wild-type PrP allele. Western blot analysis using brain homogenate from an inoculated, terminally sick  mouse. Antibodies POM1 to PrP and 4A6 to myc were used for detection. Samples were treated with PK as indicated, revealing the presence of protease resistant PrPmyc in the brain of inoculated Tg940
mouse. Antibodies POM1 to PrP and 4A6 to myc were used for detection. Samples were treated with PK as indicated, revealing the presence of protease resistant PrPmyc in the brain of inoculated Tg940  mice. (E–G) Similar neuropathological changes in hippocampus of a RML inoculated Prnp
+/o mouse and (H–K) a RML-inoculated Tg940
mice. (E–G) Similar neuropathological changes in hippocampus of a RML inoculated Prnp
+/o mouse and (H–K) a RML-inoculated Tg940  mouse. (E, H) Hematoxylin-eosin stains showing vacuolar degeneration and nerve cell loss. The dashed lines indicate the magnified area shown in F,G,J and K. Scale bar = 500 µm. (F, J) GFAP immunohistochemistry for the detection of reactive astrocytes and (G, K) mAb SAF84 for PrP aggregates. Scale bar = 200 µm. The small inserts represent the low magnification pictures of the GFAP and SAF84 stained sections consecutive to E and H. Scale bar = 500 µm.
mouse. (E, H) Hematoxylin-eosin stains showing vacuolar degeneration and nerve cell loss. The dashed lines indicate the magnified area shown in F,G,J and K. Scale bar = 500 µm. (F, J) GFAP immunohistochemistry for the detection of reactive astrocytes and (G, K) mAb SAF84 for PrP aggregates. Scale bar = 200 µm. The small inserts represent the low magnification pictures of the GFAP and SAF84 stained sections consecutive to E and H. Scale bar = 500 µm.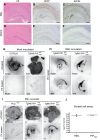
 mouse compared to (D–F) Mock inoculated Tg940
mouse compared to (D–F) Mock inoculated Tg940  mouse: (A, D) Hematoxylin-eosin stains visualizing vacuolar degeneration and nerve cell loss, (B, E) GFAP immunohistochemistry indicating reactive astrocytic gliosis, and (C, F) mAb SAF84 showing PrPmyc aggregates. Scale bar = 500 µm. (G–I) Conversion of PrPmyc into myc-tagged PK-resistant
mouse: (A, D) Hematoxylin-eosin stains visualizing vacuolar degeneration and nerve cell loss, (B, E) GFAP immunohistochemistry indicating reactive astrocytic gliosis, and (C, F) mAb SAF84 showing PrPmyc aggregates. Scale bar = 500 µm. (G–I) Conversion of PrPmyc into myc-tagged PK-resistant  in mice lacking both wild-type Prnp alleles. Histoblot analysis of coronal slices from brains of mock and prion-inoculated mice blotted onto nitrocellulose membranes. (G) Mock-inoculated C57BL/6 and Tg940
in mice lacking both wild-type Prnp alleles. Histoblot analysis of coronal slices from brains of mock and prion-inoculated mice blotted onto nitrocellulose membranes. (G) Mock-inoculated C57BL/6 and Tg940  mice. Brain homogenates were incubated with POM1 and 4A6 before or after PK treatment, and showed no PK-resistant PrP. (H) Prion-inoculated Tg940
mice. Brain homogenates were incubated with POM1 and 4A6 before or after PK treatment, and showed no PK-resistant PrP. (H) Prion-inoculated Tg940  and Tg940
and Tg940  mice, treated with PK and incubated with POM1 and 4A6, showed PK-resistant material in brain. (I) Prion-inoculated Prnp
+/o, Tg940
mice, treated with PK and incubated with POM1 and 4A6, showed PK-resistant material in brain. (I) Prion-inoculated Prnp
+/o, Tg940  and Tg940
and Tg940  mice treated with PK and untreated were stained with 4A6 anti-myc antibody and show protease-resistant PrPmyc in the brain. A terminally sick Prnp
+/o mouse was used to control for nonspecific 4A6 signals. (J) SCEPA of brain homogenates of
mice treated with PK and untreated were stained with 4A6 anti-myc antibody and show protease-resistant PrPmyc in the brain. A terminally sick Prnp
+/o mouse was used to control for nonspecific 4A6 signals. (J) SCEPA of brain homogenates of  and wild-type mouse. Three independent biological replicas of
and wild-type mouse. Three independent biological replicas of  and 2 independent biological replicas for RML were analyzed in tenfold dilution steps using 6–12 PK1-containing replica wells for each dilution. Data points indicate the number of infectious tissue culture units per ml of brain homogenates.
and 2 independent biological replicas for RML were analyzed in tenfold dilution steps using 6–12 PK1-containing replica wells for each dilution. Data points indicate the number of infectious tissue culture units per ml of brain homogenates.
 mice were used for immunoprecipitations. Specifically eluted PrPmyc protein was detected with anti-PrP antibodies as well as 4A6 anti-myc antibody (data not shown). No signal for PrP in the specific elution (s.e.) from the precipitation in 129S2/SvPas brain homogenate, and only weak signals from the elutions with unspecific peptide (u.e.), or with PBS only (b.e.). (B) Silver stain of the material immunocaptured with anti-myc antibodies from total brain homogenate of Tg940
mice were used for immunoprecipitations. Specifically eluted PrPmyc protein was detected with anti-PrP antibodies as well as 4A6 anti-myc antibody (data not shown). No signal for PrP in the specific elution (s.e.) from the precipitation in 129S2/SvPas brain homogenate, and only weak signals from the elutions with unspecific peptide (u.e.), or with PBS only (b.e.). (B) Silver stain of the material immunocaptured with anti-myc antibodies from total brain homogenate of Tg940  and eluted with myc and cym peptide. The gel was subsequently used for GeLC-MS/MS experiments. (C) Blot of peptide pair frequency against XPRESS-Ratios on a logarithmic scale. Values of ratios where one of the two labeled peptide was not detected (1∶0 or 0∶1) were excluded from the dataset. The ratio of the cystein-containing peptide pair of PrP heavy/light is indicated by the green circle. (D–F) Western blot analysis of those protein candidates listed in Table 1 for which specific antibodies were available. (D) Western blot analysis of the specific and scrambled-peptide IP elution using anti-CNPase antibody. (E–F) Western blot analyses of IP input material from wild-type 129S2/SvPas and Tg940
and eluted with myc and cym peptide. The gel was subsequently used for GeLC-MS/MS experiments. (C) Blot of peptide pair frequency against XPRESS-Ratios on a logarithmic scale. Values of ratios where one of the two labeled peptide was not detected (1∶0 or 0∶1) were excluded from the dataset. The ratio of the cystein-containing peptide pair of PrP heavy/light is indicated by the green circle. (D–F) Western blot analysis of those protein candidates listed in Table 1 for which specific antibodies were available. (D) Western blot analysis of the specific and scrambled-peptide IP elution using anti-CNPase antibody. (E–F) Western blot analyses of IP input material from wild-type 129S2/SvPas and Tg940  mice and specific and unspecific peptide elution using anti-M6 (M6-7) and anti-Neurofascin (NF155) antibodies.
mice and specific and unspecific peptide elution using anti-M6 (M6-7) and anti-Neurofascin (NF155) antibodies.References
-
- Büeler HR, Aguzzi A, Sailer A, Greiner RA, Autenried P, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. - PubMed
-
- Sailer A, Büeler H, Fischer M, Aguzzi A, Weissmann C. No propagation of prions in mice devoid of PrP. Cell. 1994;77:967–968. - PubMed
-
- Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, et al. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. - PubMed
-
- Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

