Prediction of interactions between HIV-1 and human proteins by information integration
- PMID: 19209727
- PMCID: PMC3263379
Prediction of interactions between HIV-1 and human proteins by information integration
Abstract
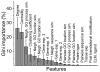
Human immunodeficiency virus-1 (HIV-1) in acquired immune deficiency syndrome (AIDS) relies on human host cell proteins in virtually every aspect of its life cycle. Knowledge of the set of interacting human and viral proteins would greatly contribute to our understanding of the mechanisms of infection and subsequently to the design of new therapeutic approaches. This work is the first attempt to predict the global set of interactions between HIV-1 and human host cellular proteins. We propose a supervised learning framework, where multiple information data sources are utilized, including co-occurrence of functional motifs and their interaction domains and protein classes, gene ontology annotations, posttranslational modifications, tissue distributions and gene expression profiles, topological properties of the human protein in the interaction network and the similarity of HIV-1 proteins to human proteins' known binding partners. We trained and tested a Random Forest (RF) classifier with this extensive feature set. The model's predictions achieved an average Mean Average Precision (MAP) score of 23%. Among the predicted interactions was for example the pair, HIV-1 protein tat and human vitamin D receptor. This interaction had recently been independently validated experimentally. The rank-ordered lists of predicted interacting pairs are a rich source for generating biological hypotheses. Amongst the novel predictions, transcription regulator activity, immune system process and macromolecular complex were the top most significant molecular function, process and cellular compartments, respectively. Supplementary material is available at URL www.cs.cmu.edu/õznur/hiv/hivPPI.html
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
