Molecular mechanism of apolipoprotein E binding to lipoprotein particles
- PMID: 19209940
- PMCID: PMC2765560
- DOI: 10.1021/bi9000694
Molecular mechanism of apolipoprotein E binding to lipoprotein particles
Abstract
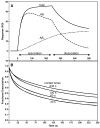
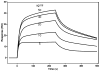
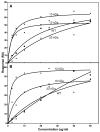
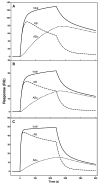
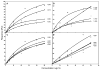
The exchangeability of apolipoprotein (apo) E between lipoprotein particles such as very low-density lipoprotein (VLDL) and high-density lipoprotein (HDL) is critical for lipoprotein metabolism, but despite its importance, the kinetics and mechanism of apoE-lipoprotein interaction are not known. We have used surface plasmon resonance (SPR) to monitor in real time the reversible binding of apoE to human VLDL and HDL(3); biotinylated lipoproteins were immobilized on a streptavidin-coated SPR sensor chip, and solutions containing various human apoE molecules at different concentrations were passed across the surface. Analysis of the resultant sensorgrams indicated that the apoE3-lipoprotein interaction is a two-step process. After an initial interaction, the second slower step involves opening of the N-terminal helix bundle domain of the apoE molecule. Destabilization of this domain leads to more rapid interfacial rearrangement which is seen when the lipoprotein binding of apoE4 is compared to that of apoE3. The resultant differences in interfacial packing seem to underlie the differing abilities of apoE4 and apoE3 to bind to VLDL and HDL(3). The measured dissociation constants for apoE binding to these lipoprotein particles are in the micromolar range. C-Terminal truncations of apoE to remove the lipid binding region spanning residues 250-299 reduce the level of binding to both types of lipoprotein, but the effect is weaker with HDL(3); this suggests that protein-protein interactions are important for apoE binding to this lipoprotein while apoE-lipid interactions are more significant for VLDL binding. The two-step mechanism of lipoprotein binding exhibited by apoE is likely to apply to other members of the exchangeable apolipoprotein family.
Figures

Similar articles
-
Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4.Biochemistry. 2010 Dec 28;49(51):10881-9. doi: 10.1021/bi1017655. Epub 2010 Dec 3. Biochemistry. 2010. PMID: 21114327 Free PMC article.
-
Contributions of the carboxyl-terminal helical segment to the self-association and lipoprotein preferences of human apolipoprotein E3 and E4 isoforms.Biochemistry. 2008 Mar 4;47(9):2968-77. doi: 10.1021/bi701923h. Epub 2008 Jan 18. Biochemistry. 2008. PMID: 18201068 Free PMC article.
-
Influence of domain stability on the properties of human apolipoprotein E3 and E4 and mouse apolipoprotein E.Biochemistry. 2014 Jun 24;53(24):4025-33. doi: 10.1021/bi500340z. Biochemistry. 2014. PMID: 24871385 Free PMC article.
-
Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders.J Mol Med (Berl). 2016 Jul;94(7):739-46. doi: 10.1007/s00109-016-1427-y. Epub 2016 Jun 9. J Mol Med (Berl). 2016. PMID: 27277824 Free PMC article. Review.
-
Apolipoprotein E isoforms and lipoprotein metabolism.IUBMB Life. 2014 Sep;66(9):616-23. doi: 10.1002/iub.1314. IUBMB Life. 2014. PMID: 25328986 Review.
Cited by
-
Neglected but Important Role of Apolipoprotein E Exchange in Hepatitis C Virus Infection.J Virol. 2016 Oct 14;90(21):9632-9643. doi: 10.1128/JVI.01353-16. Print 2016 Nov 1. J Virol. 2016. PMID: 27535051 Free PMC article.
-
ApoE: the role of conserved residues in defining function.Protein Sci. 2015 Jan;24(1):138-44. doi: 10.1002/pro.2597. Epub 2014 Dec 9. Protein Sci. 2015. PMID: 25377861 Free PMC article.
-
mRNA-based therapeutic strategies for cancer treatment.Mol Ther. 2024 Sep 4;32(9):2819-2834. doi: 10.1016/j.ymthe.2024.04.035. Epub 2024 May 3. Mol Ther. 2024. PMID: 38702886 Review.
-
New insights into the determination of HDL structure by apolipoproteins: Thematic review series: high density lipoprotein structure, function, and metabolism.J Lipid Res. 2013 Aug;54(8):2034-2048. doi: 10.1194/jlr.R034025. Epub 2012 Dec 10. J Lipid Res. 2013. PMID: 23230082 Free PMC article. Review.
-
Dissociation of apolipoprotein E oligomers to monomer is required for high-affinity binding to phospholipid vesicles.Biochemistry. 2011 Apr 5;50(13):2550-8. doi: 10.1021/bi1020106. Epub 2011 Feb 28. Biochemistry. 2011. PMID: 21322570 Free PMC article.
References
-
- Curtiss LK, Boisvert WA. Apolipoprotein E and atherosclerosis. Curr Opin Lipidol. 2000;11:243–251. - PubMed
-
- Mahley RW, Rall SCJ. Apolipoprotein E: Far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;01:507–537. - PubMed
-
- Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem. 1994;45:249–302. - PubMed
-
- Mahley RW, Ji SZ. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res. 1999;40:1–16. - PubMed
-
- Saito H, Lund-Katz S, Phillips MC. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog Lipid Res. 2004;43:350–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

