TACOA: taxonomic classification of environmental genomic fragments using a kernelized nearest neighbor approach
- PMID: 19210774
- PMCID: PMC2653487
- DOI: 10.1186/1471-2105-10-56
TACOA: taxonomic classification of environmental genomic fragments using a kernelized nearest neighbor approach
Abstract
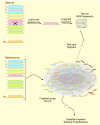
Background: Metagenomics, or the sequencing and analysis of collective genomes (metagenomes) of microorganisms isolated from an environment, promises direct access to the "unculturable majority". This emerging field offers the potential to lay solid basis on our understanding of the entire living world. However, the taxonomic classification is an essential task in the analysis of metagenomics data sets that it is still far from being solved. We present a novel strategy to predict the taxonomic origin of environmental genomic fragments. The proposed classifier combines the idea of the k-nearest neighbor with strategies from kernel-based learning.
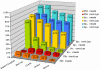
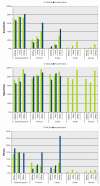
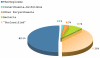
Results: Our novel strategy was extensively evaluated using the leave-one-out cross validation strategy on fragments of variable length (800 bp - 50 Kbp) from 373 completely sequenced genomes. TACOA is able to classify genomic fragments of length 800 bp and 1 Kbp with high accuracy until rank class. For longer fragments > or = 3 Kbp accurate predictions are made at even deeper taxonomic ranks (order and genus). Remarkably, TACOA also produces reliable results when the taxonomic origin of a fragment is not represented in the reference set, thus classifying such fragments to its known broader taxonomic class or simply as "unknown". We compared the classification accuracy of TACOA with the latest intrinsic classifier PhyloPythia using 63 recently published complete genomes. For fragments of length 800 bp and 1 Kbp the overall accuracy of TACOA is higher than that obtained by PhyloPythia at all taxonomic ranks. For all fragment lengths, both methods achieved comparable high specificity results up to rank class and low false negative rates are also obtained.
Conclusion: An accurate multi-class taxonomic classifier was developed for environmental genomic fragments. TACOA can predict with high reliability the taxonomic origin of genomic fragments as short as 800 bp. The proposed method is transparent, fast, accurate and the reference set can be easily updated as newly sequenced genomes become available. Moreover, the method demonstrated to be competitive when compared to the most current classifier PhyloPythia and has the advantage that it can be locally installed and the reference set can be kept up-to-date.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

