In vitro complementation of Tdp1 deficiency indicates a stabilized enzyme-DNA adduct from tyrosyl but not glycolate lesions as a consequence of the SCAN1 mutation
- PMID: 19211312
- PMCID: PMC2844109
- DOI: 10.1016/j.dnarep.2008.12.012
In vitro complementation of Tdp1 deficiency indicates a stabilized enzyme-DNA adduct from tyrosyl but not glycolate lesions as a consequence of the SCAN1 mutation
Abstract
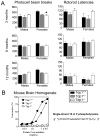
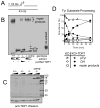
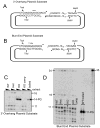
A homozygous H493R mutation in the active site of tyrosyl-DNA phosphodiesterase (TDP1) has been implicated in hereditary spinocerebellar ataxia with axonal neuropathy (SCAN1), an autosomal recessive neurodegenerative disease. However, it is uncertain how the H493R mutation elicits the specific pathologies of SCAN1. To address this question, and to further elucidate the role of TDP1 in repair of DNA end modifications and general physiology, we generated a Tdp1 knockout mouse and carried out detailed behavioral analyses as well as characterization of repair deficiencies in extracts of embryo fibroblasts from these animals. While Tdp1(-/-) mice appear phenotypically normal, extracts from Tdp1(-/-) fibroblasts exhibited deficiencies in processing 3'-phosphotyrosyl single-strand breaks and 3'-phosphoglycolate double-strand breaks (DSBs), but not 3'-phosphoglycolate single-strand breaks. Supplementing Tdp1(-/-) extracts with H493R TDP1 partially restored processing of 3'-phosphotyrosyl single-strand breaks, but with evidence of persistent covalent adducts between TDP1 and DNA, consistent with a proposed intermediate-stabilization effect of the SCAN1 mutation. However, H493R TDP1 supplementation had no effect on phosphoglycolate (PG) termini on 3' overhangs of double-strand breaks; these remained completely unprocessed. Altogether, these results suggest that for 3'-phosphoglycolate overhang lesions, the SCAN1 mutation confers loss of function, while for 3'-phosphotyrosyl lesions, the mutation uniquely stabilizes a reaction intermediate.
Figures



Similar articles
-
Deficiency in 3'-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1).Nucleic Acids Res. 2005 Jan 12;33(1):289-97. doi: 10.1093/nar/gki170. Print 2005. Nucleic Acids Res. 2005. PMID: 15647511 Free PMC article.
-
Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation?EMBO J. 2007 Nov 14;26(22):4732-43. doi: 10.1038/sj.emboj.7601885. Epub 2007 Oct 18. EMBO J. 2007. PMID: 17948061 Free PMC article.
-
Tyrosyl-DNA phosphodiesterase and the repair of 3'-phosphoglycolate-terminated DNA double-strand breaks.DNA Repair (Amst). 2009 Aug 6;8(8):901-11. doi: 10.1016/j.dnarep.2009.05.003. Epub 2009 Jun 7. DNA Repair (Amst). 2009. PMID: 19505854 Free PMC article.
-
DNA single-strand break repair and spinocerebellar ataxia with axonal neuropathy-1.Neuroscience. 2007 Apr 14;145(4):1260-6. doi: 10.1016/j.neuroscience.2006.08.048. Epub 2006 Oct 11. Neuroscience. 2007. PMID: 17045754 Review.
-
Spinocerebellar ataxia with axonal neuropathy.Adv Exp Med Biol. 2010;685:75-83. doi: 10.1007/978-1-4419-6448-9_7. Adv Exp Med Biol. 2010. PMID: 20687496 Review.
Cited by
-
Ataxia-associated DNA repair genes protect the Drosophila mushroom body and locomotor function against glutamate signaling-associated damage.Front Neural Circuits. 2023 Jul 5;17:1148947. doi: 10.3389/fncir.2023.1148947. eCollection 2023. Front Neural Circuits. 2023. PMID: 37476399 Free PMC article.
-
Role of TDP2 in the repair of DNA damage induced by the radiomimetic drug Bleomycin.Genes Environ. 2025 Mar 28;47(1):7. doi: 10.1186/s41021-025-00329-9. Genes Environ. 2025. PMID: 40155951 Free PMC article.
-
Expression profile and mitochondrial colocalization of Tdp1 in peripheral human tissues.J Mol Histol. 2013 Aug;44(4):481-94. doi: 10.1007/s10735-013-9496-5. Epub 2013 Mar 28. J Mol Histol. 2013. PMID: 23536040
-
Tyrosyl-DNA Phosphodiesterase I a critical survival factor for neuronal development and homeostasis.J Neurol Neuromedicine. 2016;1(5):25-29. doi: 10.29245/2572.942x/2016/5.1048. J Neurol Neuromedicine. 2016. PMID: 27747316 Free PMC article.
-
Topoisomerase-Mediated DNA Damage in Neurological Disorders.Front Aging Neurosci. 2021 Nov 25;13:751742. doi: 10.3389/fnagi.2021.751742. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34899270 Free PMC article. Review.
References
-
- Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. - PubMed
-
- Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. - PubMed
-
- Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, Pommier Y. DNA Repair. Vol. 5. Amst: 2006. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes; pp. 1489–1494. - PubMed
-
- Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J Biol Chem. 2002;277:27162–27168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

