MtDNA mutation pattern in tumors and human evolution are shaped by similar selective constraints
- PMID: 19211544
- PMCID: PMC2665776
- DOI: 10.1101/gr.086462.108
MtDNA mutation pattern in tumors and human evolution are shaped by similar selective constraints
Abstract
Multiple human mutational landscapes of normal and cancer conditions are currently available. However, while the unique mutational patterns of tumors have been extensively studied, little attention has been paid to similarities between malignant and normal conditions. Here we compared the pattern of mutations in the mitochondrial genomes (mtDNAs) of cancer (98 sequences) and natural populations (2400 sequences). De novo mtDNA mutations in cancer preferentially colocalized with ancient variants in human phylogeny. A significant portion of the cancer mutations was organized in recurrent combinations (COMs), reaching a length of seven mutations, which also colocalized with ancient variants. Thus, by analyzing similarities rather than differences in patterns of mtDNA mutations in tumor and human evolution, we discovered evidence for similar selective constraints, suggesting a functional potential for these mutations.
Figures
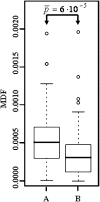

References
-
- Anderson A.R., Weaver A.M., Cummings P.T., Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127:905–915. - PubMed
-
- Bamshad M., Wooding S.P. Signatures of natural selection in the human genome. Nat. Rev. Genet. 2003;4:99–111. - PubMed
-
- Brandon M., Baldi P., Wallace D.C. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
