Differential clathrin binding and subcellular localization of OCRL1 splice isoforms
- PMID: 19211563
- PMCID: PMC2665120
- DOI: 10.1074/jbc.M807442200
Differential clathrin binding and subcellular localization of OCRL1 splice isoforms
Abstract
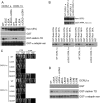
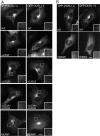
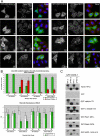
Mutation of the inositol polyphosphate 5-phosphatase OCRL1 causes the X-linked disorder oculocerebrorenal syndrome of Lowe, characterized by defects in the brain, kidneys, and eyes. OCRL1 exists as two splice isoforms that differ by a single exon encoding 8 amino acids. The longer protein, termed isoform a, is the only form in brain, whereas both isoforms are present in all other tissues. The significance of OCRL1 splicing is currently unclear. Given its proximity to a clathrin-binding site, we hypothesized that splicing may alter the clathrin binding properties of OCRL1. Here we show that this is indeed the case. OCRL1 isoform a binds clathrin with higher affinity than isoform b and is significantly more enriched in clathrin-coated trafficking intermediates. We also identify a second clathrin-binding site in OCRL1 that contributes to clathrin binding of both isoforms. Association of OCRL1 with clathrin-coated intermediates requires membrane association through interaction with Rab GTPases but not binding to the clathrin adaptor AP2. Expression of OCRL1 isoform a lacking the 5-phosphatase domain impairs transferrin endocytosis, whereas an equivalent version of isoform b does not. Our results suggest that OCRL1 exists as two functional pools, one participating in clathrin-mediated trafficking events such as endocytosis and another that is much less or not involved in this process.
Figures
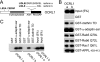




References
-
- Lowe, C. U., Terrey, M., and Mac, L. E. (1952) AMA Am. J. Dis. Child 83 164-184 - PubMed
-
- Nussbaum, R., and Suchy, S. F. (2001) in Metabolic and Molecular Basis of Inherited Diseases (Scriver, C. R., Beauder, A. L., Sly, W. S., and Valle, D., eds) pp. 6257-6266, McGraw-Hill Inc., New York
-
- Lowe, M. (2005) Traffic 6 711-719 - PubMed
-
- Faucherre, A., Desbois, P., Nagano, F., Satre, V., Lunardi, J., Gacon, G., and Dorseuil, O. (2005) Hum. Mol. Genet. 14 1441-1448 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

