Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2
- PMID: 19211564
- PMCID: PMC2665091
- DOI: 10.1074/jbc.M807365200
Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2
Abstract
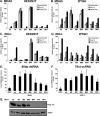
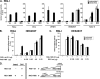
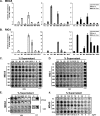
Intracellular pattern recognition receptors MDA5, RIG-I, and LGP2 are essential components of the cellular response to virus infection and are homologous to the DEXH box subfamily of RNA helicases. However, the relevance of helicase activity in the regulation of interferon production remains elusive. To examine the importance of the helicase domain function for these signaling proteins, a series of mutations targeting conserved helicase sequence motifs were analyzed for enzymatic activity, RNA binding, interferon induction, and antiviral signaling. Results indicate that all targeted motifs are required for ATP hydrolysis, but a subset is involved in RNA binding. The enzymatically inactive mutants differed in their signaling ability. Notably, mutations to MDA5 motifs I, III, and VI and RIG-I motif III produced helicase proteins with constitutive antiviral activity, whereas mutations in RIG-I motif V retained ATP hydrolysis but failed to mediate signal transduction. These findings demonstrate that type I interferon production mediated by full-length MDA5 and RIG-I is independent of the helicase domain catalytic activity. In addition, neither enzymatic activity nor RNA binding was required for negative regulation of antiviral signaling by LGP2, supporting an RNA-independent interference mechanism.
Figures

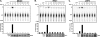


References
-
- Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783–801 - PubMed
-
- Akira, S., and Takeda, K. (2004) Nat. Rev. Immunol. 4 499–511 - PubMed
-
- Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S., and Fujita, T. (2004) Nat. Immunol. 5 730–737 - PubMed
-
- Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., Ishii, K. J., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 981–988 - PubMed
-
- Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Bartenschlager, R., and Tschopp, J. (2005) Nature 437 1167–1172 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

