Genomic epidemiology of a dengue virus epidemic in urban Singapore
- PMID: 19211734
- PMCID: PMC2668455
- DOI: 10.1128/JVI.02445-08
Genomic epidemiology of a dengue virus epidemic in urban Singapore
Abstract
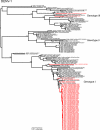
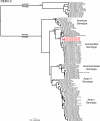
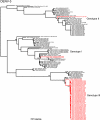
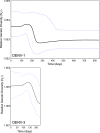
Dengue is one of the most important emerging diseases of humans, with no preventative vaccines or antiviral cures available at present. Although one-third of the world's population live at risk of infection, little is known about the pattern and dynamics of dengue virus (DENV) within outbreak situations. By exploiting genomic data from an intensively studied major outbreak, we are able to describe the molecular epidemiology of DENV at a uniquely fine-scaled temporal and spatial resolution. Two DENV serotypes (DENV-1 and DENV-3), and multiple component genotypes, spread concurrently and with similar epidemiological and evolutionary profiles during the initial outbreak phase of a major dengue epidemic that took place in Singapore during 2005. Although DENV-1 and DENV-3 differed in viremia and clinical outcome, there was no evidence for adaptive evolution before, during, or after the outbreak, indicating that ecological or immunological rather than virological factors were the key determinants of epidemic dynamics.
Figures


References
-
- Bennett, S. N., E. C. Holmes, M. Chirivella, D. M. Rodriguez, M. Beltran, V. Vorndam, D. J. Gubler, and W. O. McMillan. 2006. Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. J. Gen. Virol. 87885-893. - PubMed
-
- de Oliveira, T., O. G. Pybus, A. Rambaut, M. Salemi, S. Cassol, M. Ciccozzi, G. Rezza, G. C. Gattinara, R. D'Arrigo, M. Amicosante, L. Perrin, V. Colizzi, and C. F. Perno. 2006. Molecular epidemiology: HIV-1 and HCV sequences from Libyan outbreak. Nature 444836-837. - PubMed
-
- Drummond, A. J., A. Rambaut, B. Shapiro, and O. G. Pybus. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 221185-1192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

