JunD and HIF-1alpha mediate transcriptional activation of angiotensinogen by TGF-beta1 in human lung fibroblasts
- PMID: 19211927
- PMCID: PMC2718801
- DOI: 10.1096/fj.08-114611
JunD and HIF-1alpha mediate transcriptional activation of angiotensinogen by TGF-beta1 in human lung fibroblasts
Abstract
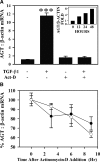
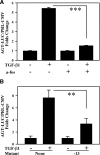
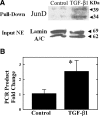
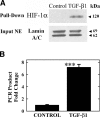
Earlier work showed that TGF-beta1 potently increases angiotensinogen (AGT) gene mRNA in primary human lung fibroblasts. Here the mechanism of TGF-beta1-induced AGT expression was studied in the IMR90 human lung fibroblast cell line. The increase in AGT mRNA induced by TGF-beta1 was completely blocked by actinomycin-D. TGF-beta1 increased the activity of a full-length human AGT promoter-luciferase reporter (AGT-LUC) but did not alter AGT mRNA half-life. Serial deletion analyses revealed that 67% of TGF-beta-inducible AGT-LUC activity resides in a small domain of the AGT core promoter; this domain contains binding sites for hypoxia-inducible factor (HIF)-1 and activation protein-1 (AP-1) transcription factors. TGF-beta1 increased HIF-1alpha protein abundance and the activity of a hypoxia-responsive element reporter; overexpression of HIF-1 increased basal AGT-LUC activity. Both oligonucleotide pulldown and chromatin immunoprecipitation assays revealed increased binding of JunD and HIF-1alpha to the AGT core promoter in response to TGF-beta1. TGF-beta1-inducible AGT-LUC was reduced by an AP-1 dominant negative or by mutation of the AP-1 site. Knockdown of either JunD or HIF-1alpha individually by siRNA partially reduced AGT-LUC. In contrast, simultaneous knockdown of both JunD and HIF-1alpha completely eliminated TGF-beta1-inducible AGT-LUC activity. These data suggest that TGF-beta1 up-regulates AGT transcription in human lung fibroblasts through a mechanism that requires both JunD and HIF-1alpha binding to the AGT core promoter. They also suggest a molecular mechanism linking hypoxia signaling and fibrogenic stimuli in the lungs.
Figures



References
-
- Weber K T, Sun Y. Recruitable ACE and tissue repair in the infarcted heart. J Renin Angiotensin Aldosterone Syst. 2000;1:295–303. - PubMed
-
- Yoshiji H, Kuriyama S, Yoshii J, Ikenaka Y, Noguchi R, Nakatani T, Tsujinoue H, Fukui H. Angiotensin-II type 1 receptor interaction is a major regulator for liver fibrosis development in rats. Hepatology. 2001;34:745–7501. - PubMed
-
- Klahr S, Morrissey J J. Comparative study of ACE inhibitors and angiotensin II receptor antagonists in interstitial scarring. Kidney Int Suppl. 1997;63:S111–S114. - PubMed
-
- Ko S H, Hong O K, Kim J W, Ahn Y B, Song K H, Cha B Y, Son H Y, Kim M J, Jeong I K, Yoon K H. High glucose increases extracellular matrix production in pancreatic stellate cells by activating the renin-angiotensin system. J Cell Biochem. 2006;98:343–355. - PubMed
-
- Marshall R P, Gohlke P, Chambers R C, Howell D C, Bottoms S E, Unger T, McAnulty R J, Laurent G J. Angiotensin II and the fibroproliferative response to acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2004;286:L156–L164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

