Detection of gene expression in an individual cell type within a cell mixture using microarray analysis
- PMID: 19212463
- PMCID: PMC2639697
- DOI: 10.1371/journal.pone.0004427
Detection of gene expression in an individual cell type within a cell mixture using microarray analysis
Abstract
Background: A central issue in the design of microarray-based analysis of global gene expression is the choice between using cells of single type and a mixture of cells. This study quantified the proportion of lipopolysaccharide (LPS) induced differentially expressed monocyte genes that could be measured in peripheral blood mononuclear cells (PBMC), and determined the extent to which gene expression in the non-monocyte cell fraction diluted or obscured fold changes that could be detected in the cell mixture.
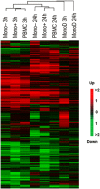
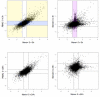
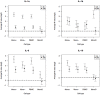
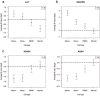
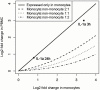
Methodology/principal findings: Human PBMC were stimulated with LPS, and monocytes were then isolated by positive (Mono+) or negative (Mono-) selection. The non-monocyte cell fraction (MonoD) remaining after positive selection of monocytes was used to determine the effect of non-monocyte cells on overall expression. RNA from LPS-stimulated PBMC, Mono+, Mono- and MonoD samples was co-hybridised with unstimulated RNA for each cell type on oligonucleotide microarrays. There was a positive correlation in gene expression between PBMC and both Mono+ (0.77) and Mono- (0.61-0.67) samples. Analysis of individual genes that were differentially expressed in Mono+ and Mono- samples showed that the ability to detect expression of some genes was similar when analysing PBMC, but for others, differential expression was either not detected or changed in the opposite direction. As a result of the dilutional or obscuring effect of gene expression in non-monocyte cells, overall about half of the statistically significant LPS-induced changes in gene expression in monocytes were not detected in PBMC. However, 97% of genes with a four fold or greater change in expression in monocytes after LPS stimulation, and almost all (96-100%) of the top 100 most differentially expressed monocyte genes were detected in PBMC.
Conclusions/significance: The effect of non-responding cells in a mixture dilutes or obscures the detection of subtle changes in gene expression in an individual cell type. However, for studies in which only the most highly differentially expressed genes are of interest, separating and analysing individual cell types may be unnecessary.
Conflict of interest statement
Figures

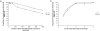
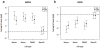

References
-
- Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, et al. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294:870–875. - PubMed
-
- Skelton L, Cooper M, Murphy M, Platt A. Human immature monocyte-derived dendritic cells express the G protein-coupled receptor GPR105 (KIAA0001, P2Y14) and increase intracellular calcium in response to its agonist, uridine diphosphoglucose. J Immunol. 2003;171:1941–1949. - PubMed
-
- Viemann D, Schulze-Osthoff K, Roth J. Potentials and pitfalls of DNA array analysis of the endothelial stress response. Biochim Biophys Acta. 2005;1746:73–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

