Disease-associated mutant ubiquitin causes proteasomal impairment and enhances the toxicity of protein aggregates
- PMID: 19214209
- PMCID: PMC2633047
- DOI: 10.1371/journal.pgen.1000382
Disease-associated mutant ubiquitin causes proteasomal impairment and enhances the toxicity of protein aggregates
Abstract
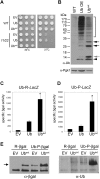
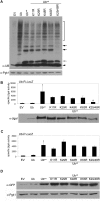
Protein homeostasis is critical for cellular survival and its dysregulation has been implicated in Alzheimer's disease (AD) and other neurodegenerative disorders. Despite the growing appreciation of the pathogenic mechanisms involved in familial forms of AD, much less is known about the sporadic cases. Aggregates found in both familial and sporadic AD often include proteins other than those typically associated with the disease. One such protein is a mutant form of ubiquitin, UBB+1, a frameshift product generated by molecular misreading of a wild-type ubiquitin gene. UBB+1 has been associated with multiple disorders. UBB+1 cannot function as a ubiquitin molecule, and it is itself a substrate for degradation by the ubiquitin/proteasome system (UPS). Accumulation of UBB+1 impairs the proteasome system and enhances toxic protein aggregation, ultimately resulting in cell death. Here, we describe a novel model system to investigate how UBB+1 impairs UPS function and whether it plays a causal role in protein aggregation. We expressed a protein analogous to UBB+1 in yeast (Ub(ext)) and demonstrated that it caused UPS impairment. Blocking ubiquitination of Ub(ext) or weakening its interactions with other ubiquitin-processing proteins reduced the UPS impairment. Expression of Ub(ext) altered the conjugation of wild-type ubiquitin to a UPS substrate. The expression of Ub(ext) markedly enhanced cellular susceptibility to toxic protein aggregates but, surprisingly, did not induce or alter nontoxic protein aggregates in yeast. Taken together, these results suggest that Ub(ext) interacts with more than one protein to elicit impairment of the UPS and affect protein aggregate toxicity. Furthermore, we suggest a model whereby chronic UPS impairment could inflict deleterious consequences on proper protein aggregate sequestration.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

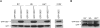
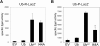



References
-
- Goedert M, Spillantini MG. A century of Alzheimer's disease. Science. 2006;314:777–781. - PubMed
-
- Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. - PubMed
-
- Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. - PubMed
-
- Schwartz AL, Ciechanover A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu Rev Med. 1999;50:57–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

