A basis for reduced chemical library inhibition of firefly luciferase obtained from directed evolution
- PMID: 19215089
- PMCID: PMC3430137
- DOI: 10.1021/jm8014525
A basis for reduced chemical library inhibition of firefly luciferase obtained from directed evolution
Abstract
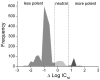
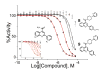
We measured the "druggability" of the ATP-dependent luciferase derived from the firefly Photuris pennsylvanica that was optimized using directed evolution (Ultra-Glo, Promega). Quantitative high-throughput screening (qHTS) was used to determine IC(50)s of 198899 samples against a formulation of Ultra-Glo luciferase (Kinase-Glo). We found that only 0.1% of the Kinase-Glo inhibitors showed an IC(50) < 10 microM compared to 0.9% found from a previous qHTS against the firefly luciferase from Photinus pyralis (lucPpy). Further, the maximum affinity identified in the lucPpy qHTS was 50 nM, while for Kinase-Glo this value increased to 600 nM. Compounds with interactions stretching outside the luciferin binding pocket were largely lost with Ultra-Glo luciferase. Therefore, Ultra-Glo luciferase will show less compound interference when used as an ATP sensor compared to lucPpy. This study demonstrates the power of large-scale quantitative analysis of structure-activity relationships (>100K compounds) in addressing important questions such as a target's druggability.
Figures

Similar articles
-
Robust light emission from cyclic alkylaminoluciferin substrates for firefly luciferase.J Am Chem Soc. 2010 Oct 6;132(39):13586-7. doi: 10.1021/ja104525m. J Am Chem Soc. 2010. PMID: 20828122 Free PMC article.
-
A Photinus pyralis and Luciola italica chimeric firefly luciferase produces enhanced bioluminescence.Biochemistry. 2014 Oct 14;53(40):6287-9. doi: 10.1021/bi501202u. Epub 2014 Oct 1. Biochemistry. 2014. PMID: 25264115
-
Quenching of the fluorescence of Tyr and Trp residues of firefly luciferase from Luciola mingrelica by the substrates.Biochemistry (Mosc). 2007 Sep;72(9):962-7. doi: 10.1134/s0006297907090064. Biochemistry (Mosc). 2007. PMID: 17922654
-
Firefly luciferase inhibition.J Photochem Photobiol B. 2010 Oct 5;101(1):1-8. doi: 10.1016/j.jphotobiol.2010.06.015. Epub 2010 Jul 3. J Photochem Photobiol B. 2010. PMID: 20655239 Review.
-
Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology.Chem Biol. 2010 Jun 25;17(6):646-57. doi: 10.1016/j.chembiol.2010.05.012. Chem Biol. 2010. PMID: 20609414 Free PMC article. Review.
Cited by
-
A homogeneous, high-throughput assay for phosphatidylinositol 5-phosphate 4-kinase with a novel, rapid substrate preparation.PLoS One. 2013;8(1):e54127. doi: 10.1371/journal.pone.0054127. Epub 2013 Jan 10. PLoS One. 2013. PMID: 23326584 Free PMC article.
-
Comprehensive characterization of cytochrome P450 isozyme selectivity across chemical libraries.Nat Biotechnol. 2009 Nov;27(11):1050-5. doi: 10.1038/nbt.1581. Epub 2009 Oct 25. Nat Biotechnol. 2009. PMID: 19855396 Free PMC article.
-
Bioluminescence in Clinical and Point-of-Care Testing.Biosensors (Basel). 2025 Jul 2;15(7):422. doi: 10.3390/bios15070422. Biosensors (Basel). 2025. PMID: 40710072 Free PMC article. Review.
-
Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter.Chem Biol. 2012 Aug 24;19(8):1060-72. doi: 10.1016/j.chembiol.2012.07.015. Chem Biol. 2012. PMID: 22921073 Free PMC article.
-
5,5-Dialkylluciferins are thermal stable substrates for bioluminescence-based detection systems.PLoS One. 2020 Dec 14;15(12):e0243747. doi: 10.1371/journal.pone.0243747. eCollection 2020. PLoS One. 2020. PMID: 33315907 Free PMC article.
References
-
- Cali JJ, Ma D, Sobol M, Simpson DJ, Frackman S, Good TD, Daily WJ, Liu D. Luminogenic cytochrome P450 assays. Expert Opin Drug Metab Toxicol. 2006;2:629–645. - PubMed
-
- Cali JJ, Niles A, Valley MP, O'Brien MA, Riss TL, Shultz J. Bioluminescent assays for ADMET. Expert Opin Drug Metab Toxicol. 2008;4:103–120. - PubMed
-
- Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol. 2007;5:127–136. - PubMed
-
- Comley J. Kinase screening and profiling-spoilt for choice. Drug Discov World. 2006;7:27–50.
-
- Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

