Functional genomics of Enterococcus faecalis: multiple novel genetic determinants for biofilm formation in the core genome
- PMID: 19218379
- PMCID: PMC2668403
- DOI: 10.1128/JB.01688-08
Functional genomics of Enterococcus faecalis: multiple novel genetic determinants for biofilm formation in the core genome
Abstract
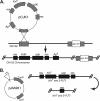
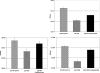
The ability of Enterococcus faecalis to form robust biofilms on host tissues and on abiotic surfaces such as catheters likely plays a major role in the pathogenesis of opportunistic antibiotic-resistant E. faecalis infections and in the transfer of antibiotic resistance genes. We have carried out a comprehensive analysis of genetic determinants of biofilm formation in the core genome of E. faecalis. Here we describe 68 genetic loci predicted to be involved in biofilm formation that were identified by recombinase in vivo expression technology (RIVET); most of these genes have not been studied previously. Differential expression of a number of these determinants during biofilm growth was confirmed by quantitative reverse transcription-PCR, and genetic complementation studies verified a role in biofilm formation for several candidate genes. Of particular interest was genetic locus EF1809, predicted to encode a regulatory protein of the GntR family. We isolated 14 independent nonsibling clones containing the putative promoter region for this gene in the RIVET screen; EF1809 also showed the largest increase in expression during biofilm growth of any of the genes tested. Since an in-frame deletion of EF1809 resulted in a severe biofilm defect that could be complemented by the cloned wild-type gene, we have designated EF1809 ebrA (enterococcal biofilm regulator). Most of the novel genetic loci identified in our studies are highly conserved in gram-positive bacterial pathogens and may thus constitute a pool of uncharacterized genes involved in biofilm formation that may be useful targets for drug discovery.
Figures


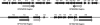
References
-
- Aravind, L., and V. Anantharaman. 2003. HutC/FarR-like bacterial transcription factors of the GntR family contain a small molecule-binding domain of the chorismate lyase fold. FEMS Microbiol. Lett. 22217-23. - PubMed
-
- Baldassarri, L., R. Creti, C. R. Arciola, L. Montanaro, M. Venditti, and R. Di Rosa. 2004. Analysis of virulence factors in cases of enterococcal endocarditis. Clin. Microbiol. Infect. 101006-1008. - PubMed
-
- Benkert, B., N. Quack, K. Schreiber, L. Jaensch, D. Jahn, and M. Schobert. 2008. Nitrate-responsive NarX-NarL represses arginine-mediated induction of the Pseudomonas aeruginosa arginine fermentation arcDABC operon. Microbiology 1543053-3060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

