Global distribution of genomic diversity underscores rich complex history of continental human populations
- PMID: 19218534
- PMCID: PMC2675968
- DOI: 10.1101/gr.088898.108
Global distribution of genomic diversity underscores rich complex history of continental human populations
Abstract
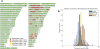
Characterizing patterns of genetic variation within and among human populations is important for understanding human evolutionary history and for careful design of medical genetic studies. Here, we analyze patterns of variation across 443,434 single nucleotide polymorphisms (SNPs) genotyped in 3845 individuals from four continental regions. This unique resource allows us to illuminate patterns of diversity in previously under-studied populations at the genome-wide scale including Latin America, South Asia, and Southern Europe. Key insights afforded by our analysis include quantifying the degree of admixture in a large collection of individuals from Guadalajara, Mexico; identifying language and geography as key determinants of population structure within India; and elucidating a north-south gradient in haplotype diversity within Europe. We also present a novel method for identifying long-range tracts of homozygosity indicative of recent common ancestry. Application of our approach suggests great variation within and among populations in the extent of homozygosity, suggesting both demographic history (such as population bottlenecks) and recent ancestry events (such as consanguinity) play an important role in patterning variation in large modern human populations.
Figures



References
-
- Barbujani G., Goldstein D. Africans and Asians abroad: Genetic diversity in Europe. Annu. Rev. Genomics Hum. Genet. 2004;5:119–150. - PubMed
-
- Bosch E., Calafell F., Perez-Lezaun A., Clarimon J., Comas D., Mateu E., Martinez-Arias R., Morera B., Brakez Z., Akhayat O., et al. Genetic structure of north-west Africa revealed by STR analysis. Eur. J. Hum. Genet. 2000;8:360–366. - PubMed
-
- Bosch E., Lee A.C., Calafell F., Arroyo E., Henneman P., de Knijff P., Jobling M.A. High resolution Y chromosome typing: 19 STRs amplified in three multiplex reactions. Forensic Sci. Int. 2002;125:42–51. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
