Characterization of a mimivirus RNA cap guanine-N2 methyltransferase
- PMID: 19218551
- PMCID: PMC2661837
- DOI: 10.1261/rna.1462109
Characterization of a mimivirus RNA cap guanine-N2 methyltransferase
Abstract
A 2,2,7-trimethylguanosine (TMG) cap is a signature feature of eukaryal snRNAs, telomerase RNAs, and trans-spliced nematode mRNAs. TMG and 2,7-dimethylguanosine (DMG) caps are also present on mRNAs of two species of alphaviruses (positive strand RNA viruses of the Togaviridae family). It is presently not known how viral mRNAs might acquire a hypermethylated cap. Mimivirus, a giant DNA virus that infects amoeba, encodes many putative enzymes and proteins implicated in RNA transactions, including the synthesis and capping of viral mRNAs and the promotion of cap-dependent translation. Here we report the identification, purification, and characterization of a mimivirus cap-specific guanine-N2 methyltransferase (MimiTgs), a monomeric enzyme that catalyzes a single round of methyl transfer from AdoMet to an m(7)G cap substrate to form a DMG cap product. MimiTgs, is apparently unable to convert a DMG cap to a TMG cap, and is thereby distinguished from the structurally homologous yeast and human Tgs1 enzymes. Nonetheless, we show genetically that MimiTgs is a true ortholog of Saccharomyces cerevisiae Tgs1. Our results hint that DMG caps can satisfy many of the functions of TMG caps in vivo. We speculate that DMG capping of mimivirus mRNAs might favor viral protein synthesis in the infected host.
Figures



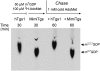

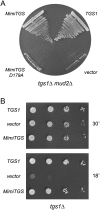
References
-
- Benarroch D., Smith P., Shuman S. Characterization of a trifunctional mimivirus mRNA capping enzyme and crystal structure of the RNA triphosphatase domain. Structure. 2008;16:501–512. - PubMed
-
- Busch H., Reddy R., Rothblum L., Choi Y.C. SnRNAs, SnRNPs, and RNA processing. Annu. Rev. Biochem. 1982;51:617–654. - PubMed
-
- Cai A., Jankowska-Anyszka M., Centers A., Chlebicka L., Stepinki J., Stolarski R., Darzynkiewicz E., Rhoads R.E. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry. 1999;38:8538–8547. - PubMed
-
- Claverie J.M., Ogata H., Audic S., Abergel C., Suhre K., Fournier P.E. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006;117:133–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
