Histone arginine methylations: their roles in chromatin dynamics and transcriptional regulation
- PMID: 19220199
- PMCID: PMC5433800
- DOI: 10.1042/BSR20080176
Histone arginine methylations: their roles in chromatin dynamics and transcriptional regulation
Abstract
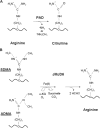
PRMTs (protein arginine N-methyltransferases) specifically modify the arginine residues of key cellular and nuclear proteins as well as histone substrates. Like lysine methylation, transcriptional repression or activation is dependent upon the site and type of arginine methylation on histone tails. Recent discoveries imply that histone arginine methylation is an important modulator of dynamic chromatin regulation and transcriptional controls. However, under the shadow of lysine methylation, the roles of histone arginine methylation have been under-explored. The present review focuses on the roles of histone arginine methylation in the regulation of gene expression, and the interplays between histone arginine methylation, histone acetylation, lysine methylation and chromatin remodelling factors. In addition, we discuss the dynamic regulation of arginine methylation by arginine demethylases, and how dysregulation of PRMTs and their activities are linked to human diseases such as cancer.
Figures






References
-
- Bedford MT, Richard S. Arginine methylation: an emerging regulator of protein function. Mol Cell. 2005;18:263–272. - PubMed
-
- Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, Pestka S. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2006;113:50–87. - PubMed
-
- McBride AE, Silver PA. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. - PubMed
-
- Cook JR, Lee JH, Yang ZH, Krause CD, Herth N, Hoffmann R, Pestka S. FBXO11/PRMT9 a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem Biophys Res Commun. 2006;342:472–481. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

