Diatom plastids depend on nucleotide import from the cytosol
- PMID: 19221027
- PMCID: PMC2642474
- DOI: 10.1073/pnas.0808862106
Diatom plastids depend on nucleotide import from the cytosol
Abstract
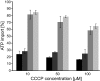
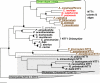
Diatoms are ecologically important algae that acquired their plastids by secondary endosymbiosis, resulting in a more complex cell structure and an altered distribution of metabolic pathways when compared with organisms with primary plastids. Diatom plastids are surrounded by 4 membranes; the outermost membrane is continuous with the endoplasmic reticulum. Genome analyses suggest that nucleotide biosynthesis is, in contrast to higher plants, not located in the plastid, but in the cytosol. As a consequence, nucleotides have to be imported into the organelle. However, the mechanism of nucleotide entry into the complex plastid is unknown. We identified a high number of putative nucleotide transporters (NTTs) in the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum and characterized the first 2 isoforms (NTT1 and NTT2). GFP-based localization studies revealed that both investigated NTTs are targeted to the plastid membranes, and that NTT1 most likely enters the innermost plastid envelope via the stroma. Heterologously expressed NTT1 acts as a proton-dependent adenine nucleotide importer, whereas NTT2 facilitates the counter exchange of (deoxy-)nucleoside triphosphates. Therefore, these transporters functionally resemble NTTs from obligate intracellular bacteria with an impaired nucleotide metabolism rather than ATP/ADP exchanging NTTs from primary plastids. We suggest that diatoms harbor a specifically-adapted nucleotide transport system and that NTTs are the key players in nucleotide supply to the complex plastid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
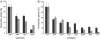

References
-
- Falkowski PG, et al. The evolution of modern eukaryotic phytoplankton. Science. 2004;305:354–360. - PubMed
-
- Roberts K, Granum E, Leegood RC, Raven JA. Carbon acquisition by diatoms. Photosynth Res. 2007;93:79–88. - PubMed
-
- Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306:79–86. - PubMed
-
- Delwiche CF. Tracing the thread of plastid diversity through the tapestry of life. Am Nat. 1999;154:S164–S177. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

