nELAV proteins alteration in Alzheimer's disease brain: a novel putative target for amyloid-beta reverberating on AbetaPP processing
- PMID: 19221430
- PMCID: PMC6057145
- DOI: 10.3233/JAD-2009-0967
nELAV proteins alteration in Alzheimer's disease brain: a novel putative target for amyloid-beta reverberating on AbetaPP processing
Abstract
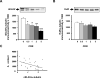
Neuronal ELAV (nELAV) proteins are RNA-binding proteins which play a physiological role in controlling gene expression in memory formation, and their alteration may contribute to cognitive impairment associated with neurodegenerative pathologies such as Alzheimer's disease (AD). Indeed, we found that the content of nELAV proteins is significantly decreased along with clinical dementia progression in the hippocampi of AD brains, where it inversely correlates with the amount of amyloid-beta (Abeta). To check the direct influence of Abeta on nELAV, we performed in vitro experiments using human SH-SY5Y cells, finding that Abeta(1-42) specifically determines nELAV proteins reduction. Since ADAM10 mRNA has the predicted sequences targeted by nELAV, we investigated whether Abeta, through nELAV proteins, could originate a vicious circle affecting amyloid-beta protein precursor (AbetaPP) processing. Immunoprecipitation experiments showed that indeed nELAV proteins bind to ADAM10 mRNA and that this binding is disrupted by Abeta(1-42) exposure, resulting in a decreased ADAM10 protein expression. ADAM10 protein diminution was also found in AD hippocampi. These data show for the first time the involvement of nELAV in AD pathology and suggest that their alteration may affect genes implicated in AbetaPP processing.
Figures


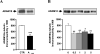
References
-
- Perrone-Bizzozero N, Bolognani F. Role of HuD and other RNA-binding proteins in neural development and plasticity. J Neurosci Res. 2002;68:121–126. - PubMed
-
- Pascale A, Gusev PA, Amadio M, Dottorini T, Govoni S, Alkon DL, Quattrone A. Neuronal ELAV proteins enhance mRNA stability by a PKC alpha-dependent pathway. Proc Natl Acad Sci USA. 2004;101:1217–1222. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

