Translational regulation of GluR2 mRNAs in rat hippocampus by alternative 3' untranslated regions
- PMID: 19222700
- PMCID: PMC2727682
- DOI: 10.1111/j.1471-4159.2009.05992.x
Translational regulation of GluR2 mRNAs in rat hippocampus by alternative 3' untranslated regions
Abstract
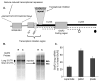
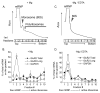
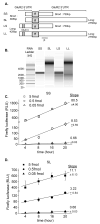
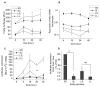
The glutamate receptor 2 (GluR2) subunit determines many of the functional properties of the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionate subtype of glutamate receptor. The roles of untranslated regions (UTRs) in mRNA stability, transport, or translation are increasingly recognized. The 3' end of the GluR2 transcripts are alternatively processed to form a short and long 3'UTR, giving rise to two pools of GluR2 mRNA of 4 and 6 kb in length, respectively, in the mammalian brain. However, the role of these alternative 3'UTRs in GluR2 expression has not been reported. We demonstrate that in the cytoplasm of rat hippocampus, native GluR2 mRNAs bearing the long 3'UTR are mostly retained in translationally dormant complexes of ribosome-free messenger ribonucleoprotein (mRNP), whereas GluR2 transcripts bearing the short 3'UTR are predominantly associated with actively translating ribosomes. One day after pilocarpine-induced status epilepticus (SE), the levels of both long and short GluR2 transcripts were markedly decreased in rat hippocampus. However, GluR2 mRNAs bearing the long 3'-UTRs were shifted from untranslating mRNP complexes to ribosome-containing complexes after SE, pointing to a selective translational derepression of GluR2 mRNA mediated by the long 3'UTR. In Xenopus oocytes, expression of firefly luciferase reporters bearing alternative GluR2 3'UTRs confirmed that the long 3'UTR is sufficient to suppress translation. The stability of reporter mRNAs in oocytes was not significantly influenced by alternative 5' or 3'UTRs of GluR2 over the time period examined. Overall, our findings that the long 3'UTR of GluR2 mRNA alone is sufficient to suppress translation, and the evidence for seizure-induced derepression of translation of GluR2 via the long 3'UTR strongly suggests that a regulatory signaling mechanism exists that differentially targets GluR2 transcripts with alternative 3'UTRs.
Figures

Similar articles
-
Control of glutamate receptor 2 (GluR2) translational initiation by its alternative 3' untranslated regions.Mol Pharmacol. 2009 Dec;76(6):1145-9. doi: 10.1124/mol.109.060343. Epub 2009 Sep 30. Mol Pharmacol. 2009. PMID: 19794129 Free PMC article.
-
Distinct 3'UTRs differentially regulate activity-dependent translation of brain-derived neurotrophic factor (BDNF).Proc Natl Acad Sci U S A. 2010 Sep 7;107(36):15945-50. doi: 10.1073/pnas.1002929107. Epub 2010 Aug 23. Proc Natl Acad Sci U S A. 2010. PMID: 20733072 Free PMC article.
-
Analyses of zebrafish and Xenopus oocyte maturation reveal conserved and diverged features of translational regulation of maternal cyclin B1 mRNA.BMC Dev Biol. 2009 Jan 28;9:7. doi: 10.1186/1471-213X-9-7. BMC Dev Biol. 2009. PMID: 19175933 Free PMC article.
-
3'UTR Diversity: Expanding Repertoire of RNA Alterations in Human mRNAs.Mol Cells. 2023 Jan 31;46(1):48-56. doi: 10.14348/molcells.2023.0003. Epub 2023 Jan 20. Mol Cells. 2023. PMID: 36697237 Free PMC article. Review.
-
3'UTRs take a long shot in the brain.Bioessays. 2014 Jan;36(1):39-45. doi: 10.1002/bies.201300100. Epub 2013 Sep 20. Bioessays. 2014. PMID: 24115048 Free PMC article. Review.
Cited by
-
Alternative polyadenylation in glioblastoma multiforme and changes in predicted RNA binding protein profiles.OMICS. 2013 Mar;17(3):136-49. doi: 10.1089/omi.2012.0098. Epub 2013 Feb 19. OMICS. 2013. PMID: 23421905 Free PMC article.
-
Control of glutamate receptor 2 (GluR2) translational initiation by its alternative 3' untranslated regions.Mol Pharmacol. 2009 Dec;76(6):1145-9. doi: 10.1124/mol.109.060343. Epub 2009 Sep 30. Mol Pharmacol. 2009. PMID: 19794129 Free PMC article.
-
Splicing and editing of ionotropic glutamate receptors: a comprehensive analysis based on human RNA-Seq data.Cell Mol Life Sci. 2021 Jul;78(14):5605-5630. doi: 10.1007/s00018-021-03865-z. Epub 2021 Jun 8. Cell Mol Life Sci. 2021. PMID: 34100982 Free PMC article.
-
Glutamate receptor ion channels: structure, regulation, and function.Pharmacol Rev. 2010 Sep;62(3):405-96. doi: 10.1124/pr.109.002451. Pharmacol Rev. 2010. PMID: 20716669 Free PMC article. Review.
-
The Rpe65 rd12 allele exerts a semidominant negative effect on vision in mice.Invest Ophthalmol Vis Sci. 2014 Apr 17;55(4):2500-15. doi: 10.1167/iovs.13-13574. Invest Ophthalmol Vis Sci. 2014. PMID: 24644049 Free PMC article.
References
-
- Awobuluyi M, Lipton SA, Sucher NJ. Translationally distinct populations of NMDA receptor subunit NR1 mRNA in the developing rat brain. J Neurochem. 2003a;87(5):1066–1075. - PubMed
-
- Awobuluyi M, Vazhappilly R, Sucher NJ. Translational activity of N-methyl-D-aspartate receptor subunit NR1 mRNA in PC12 cells. Neuro-Signals. 2003b;12(6):283–291. - PubMed
-
- Colbourne F, Grooms SY, Zukin RS, Buchan AM, Bennett MV. Hypothermia rescues hippocampal CA1 neurons and attenuates down-regulation of the AMPA receptor GluR2 subunit after forebrain ischemia. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2906–2910. - PMC - PubMed
-
- Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nature medicine. 2000;6(6):637–641. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

