KinImmerse: Macromolecular VR for NMR ensembles
- PMID: 19222844
- PMCID: PMC2650690
- DOI: 10.1186/1751-0473-4-3
KinImmerse: Macromolecular VR for NMR ensembles
Abstract
Background: In molecular applications, virtual reality (VR) and immersive virtual environments have generally been used and valued for the visual and interactive experience - to enhance intuition and communicate excitement - rather than as part of the actual research process. In contrast, this work develops a software infrastructure for research use and illustrates such use on a specific case.
Methods: The Syzygy open-source toolkit for VR software was used to write the KinImmerse program, which translates the molecular capabilities of the kinemage graphics format into software for display and manipulation in the DiVE (Duke immersive Virtual Environment) or other VR system. KinImmerse is supported by the flexible display construction and editing features in the KiNG kinemage viewer and it implements new forms of user interaction in the DiVE.
Results: In addition to molecular visualizations and navigation, KinImmerse provides a set of research tools for manipulation, identification, co-centering of multiple models, free-form 3D annotation, and output of results. The molecular research test case analyzes the local neighborhood around an individual atom within an ensemble of nuclear magnetic resonance (NMR) models, enabling immersive visual comparison of the local conformation with the local NMR experimental data, including target curves for residual dipolar couplings (RDCs).
Conclusion: The promise of KinImmerse for production-level molecular research in the DiVE is shown by the locally co-centered RDC visualization developed there, which gave new insights now being pursued in wider data analysis.
Figures

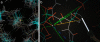

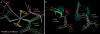
References
-
- Richardson DC, Richardson JS. Teaching molecular 3-D literacy. Biochem Molec Biol Educ. 2002;30:21–26. doi: 10.1002/bmb.2002.494030010005. - DOI
-
- Katz L, Levinthal C. Interactive computer graphics and representation of complex biological structures. Ann Rev Biophys Bioengin. 1972;1:465–504. - PubMed
-
- Porter TK. Spherical shading. Computer Graphics. 1978;12:282–285.
-
- Britton EG, Lipscomb JL, Pique ME. Making nested rotations convenient for the user. Computer Graphics. 1978;12:222–227.
-
- Jones TA. A graphics model building and refinement system for macromolecules. J Applied Crystallogr. 1978;11:268–272.
Grants and funding
LinkOut - more resources
Full Text Sources

