The structural dynamics of macromolecular processes
- PMID: 19223165
- PMCID: PMC2774249
- DOI: 10.1016/j.ceb.2009.01.022
The structural dynamics of macromolecular processes
Abstract
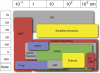
Dynamic processes involving macromolecular complexes are essential to cell function. These processes take place over a wide variety of length scales from nanometers to micrometers, and over time scales from nanoseconds to minutes. As a result, information from a variety of different experimental and computational approaches is required. We review the relevant sources of information and introduce a framework for integrating the data to produce representations of dynamic processes.
Figures


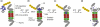
References
-
- Sali A, Chiu W. Macromolecular Assemblies Highlighted. Structure. 2005;13:339–341.
-
- van Oijen AM. Single-molecule studies of complex systems: the replisome. Mol Biosyst. 2007;3:117–125. - PubMed
-
- Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annu Rev Biochem. 2008;77:177–203. - PubMed
-
A review of single molecule experiments used to study translation in the ribosome.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

