Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing
- PMID: 19223322
- PMCID: PMC2665243
- DOI: 10.1093/nar/gkp080
Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing
Abstract
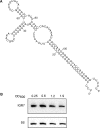
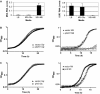
Direct cloning and parallel sequencing, an extremely powerful method for microRNA (miRNA) discovery, has not yet been applied to bacterial transcriptomes. Here we present sRNA-Seq, an unbiased method that allows for interrogation of the entire small, non-coding RNA (sRNA) repertoire in any prokaryotic or eukaryotic organism. This method includes a novel treatment that depletes total RNA fractions of highly abundant tRNAs and small subunit rRNA, thereby enriching the starting pool for sRNA transcripts with novel functionality. As a proof-of-principle, we applied sRNA-Seq to the human pathogen Vibrio cholerae. Our results provide information, at unprecedented depth, on the complexity of the sRNA component of a bacterial transcriptome. From 407 039 sequence reads, all 20 known V. cholerae sRNAs, 500 new, putative intergenic sRNAs and 127 putative antisense sRNAs were identified in a limited number of growth conditions examined. In addition, characterization of a subset of the newly identified transcripts led to the identification of a novel sRNA regulator of carbon metabolism. Collectively, these results strongly suggest that the number of sRNAs in bacteria has been greatly underestimated and that future efforts to analyze bacterial transcriptomes will benefit from direct cloning and parallel sequencing experiments aided by 5S/tRNA depletion.
Figures


Similar articles
-
Discovery of bacterial sRNAs by high-throughput sequencing.Methods Mol Biol. 2011;733:63-79. doi: 10.1007/978-1-61779-089-8_5. Methods Mol Biol. 2011. PMID: 21431763 Free PMC article.
-
Transcriptomic Approaches for Studying Quorum Sensing in Vibrio cholerae.Methods Enzymol. 2018;612:303-342. doi: 10.1016/bs.mie.2018.09.008. Epub 2018 Oct 24. Methods Enzymol. 2018. PMID: 30502947
-
Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2.Nucleic Acids Res. 2006;34(12):3484-93. doi: 10.1093/nar/gkl453. Nucleic Acids Res. 2006. PMID: 16870723 Free PMC article.
-
Identification of bacterial small non-coding RNAs: experimental approaches.Curr Opin Microbiol. 2007 Jun;10(3):257-61. doi: 10.1016/j.mib.2007.05.003. Epub 2007 Jun 5. Curr Opin Microbiol. 2007. PMID: 17553733 Review.
-
Non-coding sRNAs regulate virulence in the bacterial pathogen Vibrio cholerae.RNA Biol. 2012 Apr;9(4):392-401. doi: 10.4161/rna.19975. Epub 2012 Apr 1. RNA Biol. 2012. PMID: 22546941 Free PMC article. Review.
Cited by
-
Mycobacterial RNA isolation optimized for non-coding RNA: high fidelity isolation of 5S rRNA from Mycobacterium bovis BCG reveals novel post-transcriptional processing and a complete spectrum of modified ribonucleosides.Nucleic Acids Res. 2015 Mar 11;43(5):e32. doi: 10.1093/nar/gku1317. Epub 2014 Dec 24. Nucleic Acids Res. 2015. PMID: 25539917 Free PMC article.
-
Comparative profiling of Pseudomonas aeruginosa strains reveals differential expression of novel unique and conserved small RNAs.PLoS One. 2012;7(5):e36553. doi: 10.1371/journal.pone.0036553. Epub 2012 May 10. PLoS One. 2012. PMID: 22590564 Free PMC article.
-
How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes?BMC Genomics. 2012 Dec 27;13:734. doi: 10.1186/1471-2164-13-734. BMC Genomics. 2012. PMID: 23270466 Free PMC article.
-
A combination of improved differential and global RNA-seq reveals pervasive transcription initiation and events in all stages of the life-cycle of functional RNAs in Propionibacterium acnes, a major contributor to wide-spread human disease.BMC Genomics. 2013 Sep 14;14:620. doi: 10.1186/1471-2164-14-620. BMC Genomics. 2013. PMID: 24034785 Free PMC article.
-
Global discovery of small RNAs in Yersinia pseudotuberculosis identifies Yersinia-specific small, noncoding RNAs required for virulence.Proc Natl Acad Sci U S A. 2011 Sep 13;108(37):E709-17. doi: 10.1073/pnas.1101655108. Epub 2011 Aug 29. Proc Natl Acad Sci U S A. 2011. PMID: 21876162 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

