The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication
- PMID: 19223333
- PMCID: PMC2673421
- DOI: 10.1093/nar/gkp065
The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication
Abstract
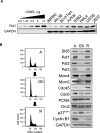
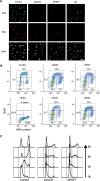
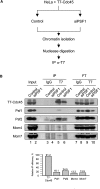
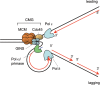
The GINS complex, originally discovered in Saccharomyces cerevisiae and Xenopus laevis, binds to DNA replication origins shortly before the onset of S phase and travels with the replication forks after initiation. In this study we present a detailed characterization of the human GINS (hGINS) homolog. Using new antibodies that allow the detection of endogenous hGINS in cells and tissues, we have examined its expression, abundance, subcellular localization and association with other DNA replication proteins. Expression of hGINS is restricted to actively proliferating cells. During the S phase, hGINS becomes part of a Cdc45-MCM-GINS (CMG) complex that is assembled on chromatin. Down-regulation of hGINS destabilizes CMG, causes a G1-S arrest and slows down ongoing DNA replication, effectively blocking cell proliferation. Our data support the notion that hGINS is an essential component of the human replisome.
Figures



References
-
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. - PubMed
-
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. - PubMed
-
- Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423:720–724. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

